I am trying to make a statistical model summary table using the modelsummary package.
I ran 6 regressions. I conducted joint hypothesis tests for 2 of them, which include certain variables (specificaly, educ and exper) as explanatory variables. I want to include the F statistics from those tests in the table, but have trouble extracting information from the tests and importing it into the table.
Here are some more details.
(1) What I did, part 1: Packages, data, and analyses
library(tidyverse)
library(wooldridge) # Data
library(estimatr) # Estimation
library(car) # F test
library(modelsummary) # Table
library(kableExtra) # Table
wage2 <- wage2 %>%
mutate(lwage = log(wage))
model1 <- lm_robust(lwage ~ educ,
data = wage2, se_type = "HC1")
model2 <- lm_robust(lwage ~ exper,
data = wage2, se_type = "HC1")
model3 <- lm_robust(lwage ~ educ exper,
data = wage2, se_type = "HC1")
F3 <- linearHypothesis(model3, test = "F",
c("educ = 0", "exper = 0"))
model4 <- lm_robust(lwage ~ educ tenure,
data = wage2, se_type = "HC1")
model5 <- lm_robust(lwage ~ exper tenure,
data = wage2, se_type = "HC1")
model6 <- lm_robust(lwage ~ educ exper tenure,
data = wage2, se_type = "HC1")
F6 <- linearHypothesis(model6, test = "F",
c("educ = 0", "exper = 0"))
The results of the joint hypothesis tests are as follows.
F3
## Linear hypothesis test
##
## Hypothesis:
## educ = 0
## exper = 0
##
## Model 1: restricted model
## Model 2: lwage ~ educ exper
##
## Res.Df Df F Pr(>F)
## 1 934
## 2 932 2 67.715 < 2.2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
F6
## Linear hypothesis test
##
## Hypothesis:
## educ = 0
## exper = 0
##
## Model 1: restricted model
## Model 2: lwage ~ educ exper tenure
##
## Res.Df Df F Pr(>F)
## 1 933
## 2 931 2 63.592 < 2.2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(2) What I did, part 2: Table
regs <- list()
regs[['Model 1']] <- model1
regs[['Model 2']] <- model2
regs[['Model 3']] <- model3
regs[['Model 4']] <- model4
regs[['Model 5']] <- model5
regs[['Model 6']] <- model6
gm <- tribble(
~raw, ~clean, ~fmt,
"adj.r.squared", "$\\bar{R}^2$", 3,
"nobs", "Sample size", 0
)
rows <- tribble(
~term, ~'Model 1', ~'Model 2', ~'Model 3',
~'Model 4', ~'Model 5', ~'Model 6',
'$F$ statstics', '', '', '', '', '', '',
'$H_0: \\beta_{\\rm{educ}} = 0, \\beta_{\\rm{exper}} = 0$',
'', '', '67.715***', '', '', '63.592***',
'', '', '', '(0.000)', '', '', '(0.000)'
)
attr(rows, 'position') <- c(9:11)
table <- msummary(regs,
estimate = "{estimate}{stars}",
gof_map = gm,
notes = list("*** p < 0.01, ** p < 0.05, * p < 0.1.
Heteroskedasticity-consistent
standard errors in brackets.
P values in brackets for F statistics."),
add_rows = rows) %>%
row_spec(c(1, 3, 5, 7, 9, 10),
extra_css = "border-bottom: hidden")
table
The above code gave me a nice table. (Unfortunately, I do not seem to be able to post it because I have not earned enough reputation.)
(3) What I want to know
In the above, I manually typed in the F statistics, stars, and p-values in rows to obtain the table I wanted, but I would like to find out if there is a way to extract information from F3 and F6 and import it to the table.
CodePudding user response:
If I understand correctly, you want to:
- Add two rows to the bottom of the table: F statistic and associated p-value
- These rows should be added only to some models, which you pick manually
The simplest way to achieve this is to use the add_rows argument you use in your example. Your approach is correct!
If you want a more general and automated strategy, 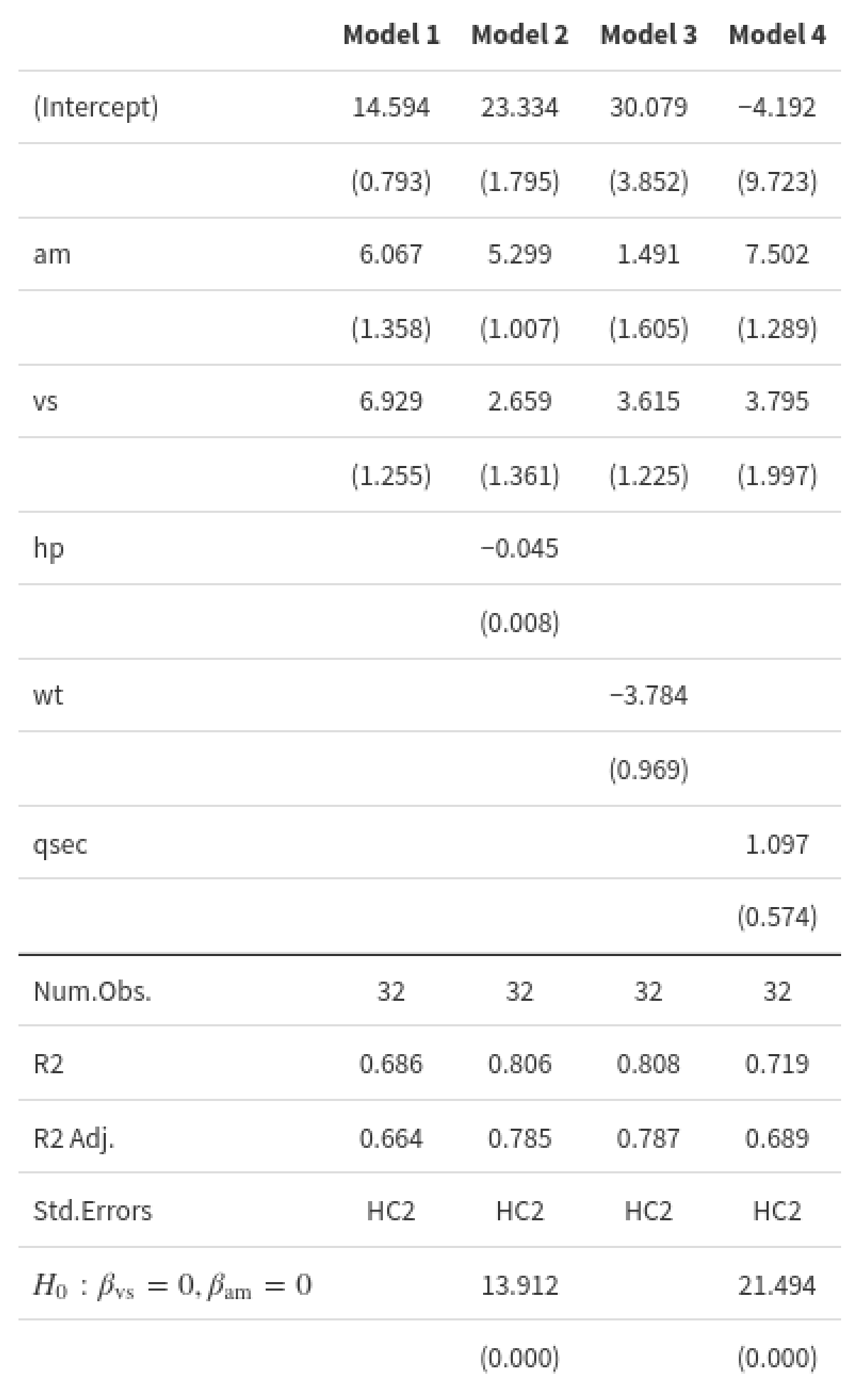
CodePudding user response:
If I understand correctly, you would like to export data from the model. Just call it out:
F3$F
and
F3$`Pr(>F)`
