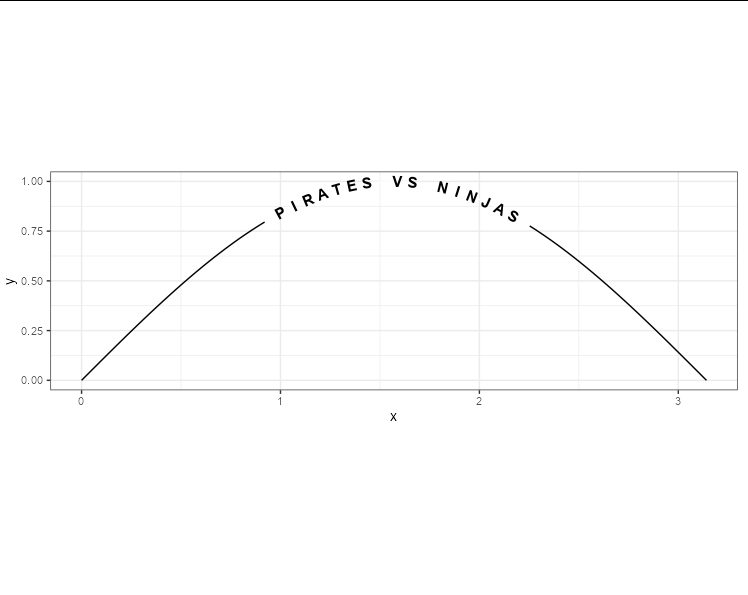
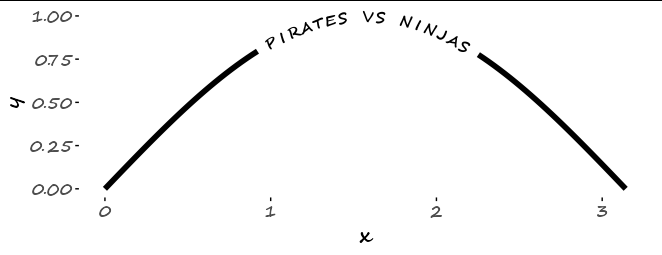
Is there a way to put text along a density line, or for that matter, any path, in ggplot2? By that, I mean either once as a label, in this style of xkcd: 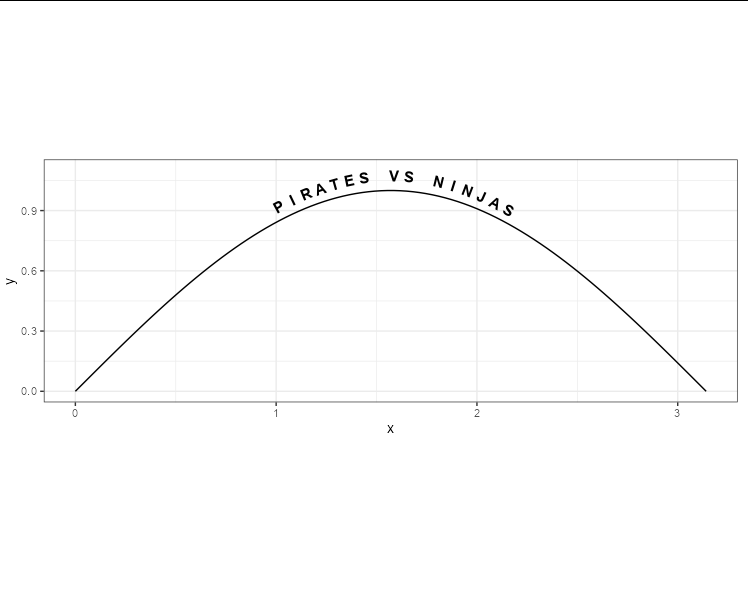
Or, if we want to replace part of the line with text:
df$col <- cut(df$x, c(-1, 0.95, 2.24, 5), c("black", "white", "#000000"))
ggplot(df, aes(x, y))
geom_line(aes(color = col, group = col))
geom_text(aes(label = label, angle = angle), data = df2,
size = 4, fontface = "bold")
scale_color_identity()
coord_equal()
theme_bw()
or, with some theme tweaks:
Addendum
Realistically, I probably won't get round to writing a geom_textpath package, but I thought it would be useful to show the sort of approach that might work for labelling density curves as per the OP's example. It requires the following suite of functions:
gradient_to_text_angle <- function(grad, mult = 1)
{
angle <- mult * atan(grad) * 180 / pi
}
get_path_data <- function(x, y)
{
grad <- diff(y)/diff(x)
multiplier <- diff(range(x))/diff(range(y))
new_x <- (head(x, -1) tail(x, -1)) / 2
new_y <- (head(y, -1) tail(y, -1)) / 2
path_length <- cumsum(sqrt(diff(x)^2 diff(multiplier * y)^2))
data.frame(x = new_x, y = new_y, gradient = grad,
angle = gradient_to_text_angle(grad, multiplier/5),
length = path_length)
}
get_path_points <- function(path, x_start, x_end, n)
{
start_dist <- approx(x = path$x, y = path$length, xout = x_start)$y
end_dist <- approx(x = path$x, y = path$length, xout = x_end)$y
dist_points <- seq(start_dist, end_dist, length.out = n)
x <- approx(x = path$length, y = path$x, xout = dist_points)$y
y <- approx(x = path$length, y = path$y, xout = dist_points)$y
grad <- approx(x = path$length, y = path$gradient, xout = dist_points)$y
angle <- approx(x = path$length, y = path$angle, xout = dist_points)$y
data.frame(x = x, y = y, gradient = grad,
angle = angle, length = dist_points)
}
label_to_path <- function(label, path, x_start = head(path$x, 1),
x_end = tail(path$x, 1))
{
letters <- unlist(strsplit(label, "")[1])
df <- get_path_points(path, x_start, x_end, length(letters))
df$letter <- letters
df
}
get_densities <- function(var, groups)
{
if(missing(groups)) values <- list(var)
else values <- split(var, groups)
lapply(values, function(x) {
d <- density(x)
data.frame(x = d$x, y = d$y)})
}
density_labels <- function(var, groups, proportion = 0.25)
{
d <- get_densities(var, groups)
d <- lapply(d, function(x) get_path_data(x$x, x$y))
labels <- unique(groups)
x_starts <- lapply(d, function(x) x$x[round((length(x$x) * (1 - proportion))/2)])
x_ends <- lapply(d, function(x) x$x[round((length(x$x) * (1 proportion))/2)])
do.call(rbind, lapply(seq_along(d), function(i) {
df <- label_to_path(labels[i], d[[i]], x_starts[[i]], x_ends[[i]])
df$group <- labels[i]
df}))
}
With these functions defined, we can now do:
set.seed(100)
df <- data.frame(value = rpois(100, 3),
group = rep(paste("This is a nice long label",
"that will demonstrate the ability",
"of text to follow a density curve"), 100))
ggplot(df, aes(value))
geom_text(aes(x = x, y = y, label = letter, angle = angle),
data = density_labels(df$value, df$group, 0.8))
theme(legend.position = "none")
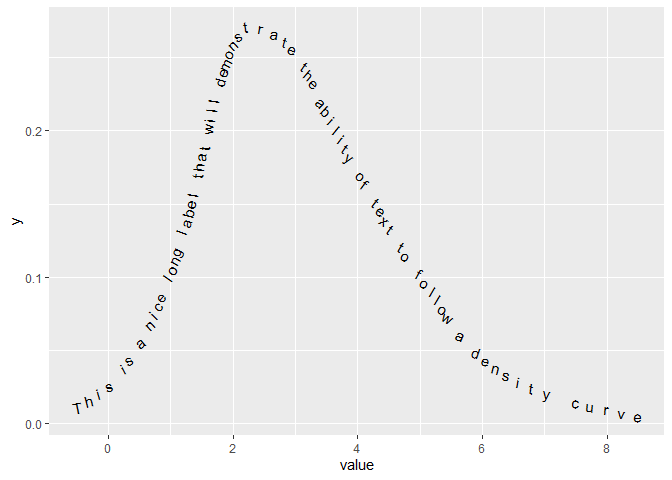
Created on 2021-11-07 by the reprex package (v2.0.0)