Here is a section of my original data frame which is survey data.
structure(list(Ages = c(30L, 30L, 30L, 30L, 30L, 33L, 33L, 27L,
27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L,
27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L,
27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L,
27L, 27L, 27L, 27L, 28L, 28L, 25L, 25L, 25L, 25L, 25L, 25L, 25L,
25L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L,
29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 38L,
38L, 38L, NA, NA, NA, NA, 31L, 31L, 31L, 31L, 31L, 31L, 33L,
33L, 33L, 33L, 33L, 33L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L,
29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L,
29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L, 29L,
29L, 29L, 34L, 34L, 34L, 34L, 34L, 34L, 34L, 34L, 34L, 34L, 34L,
34L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L,
27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L, 36L, 36L,
36L, 36L, 28L, 28L, 28L, 28L, 28L, 28L, 28L, 28L, 28L, 28L, 32L,
32L, 32L, 32L, 32L, 32L, 32L, 32L, 32L, 32L, 32L, 32L, 32L, 32L,
32L, 32L, 32L, 32L, 32L, 32L, 28L, 28L, 28L, 28L, 27L, 27L, 27L,
27L, 27L, 27L, 27L, 27L, 32L, 32L, 32L, 32L, 32L, 32L, 32L, 32L,
32L, 30L, 30L, 30L, 30L, 26L, 27L, 27L, 27L, 27L, 27L, 27L, 27L,
27L, 27L, 27L, 27L, 27L, 27L, 27L, 27L), value = c("Response Eight",
"Response Twelve", "Response Eleven", "Response Three", "Response Two",
"Response Seven", "Response Seven", "Response Eight", "Response Nine",
"Response Twelve", "Response Eleven", "Response Three", "Response Ten",
"Response Two", "Response One", "Response Four", "Response Five",
"Response Six", "Response Eight", "Response Nine", "Response Twelve",
"Response Eleven", "Response Three", "Response Ten", "Response Two",
"Response One", "Response Four", "Response Five", "Response Six",
"Response Eight", "Response Nine", "Response Twelve", "Response Eleven",
"Response Three", "Response Ten", "Response Two", "Response One",
"Response Four", "Response Five", "Response Six", "Response Eight",
"Response Nine", "Response Twelve", "Response Eleven", "Response Three",
"Response Ten", "Response Two", "Response One", "Response Four",
"Response Five", "Response Six", "Response Seven", "Response Seven",
"Response Three", "Response One", "Response Three", "Response One",
"Response Three", "Response One", "Response Three", "Response One",
"Response Twelve", "Response Three", "Response Twelve", "Response Three",
"Response Twelve", "Response Three", "Response Twelve", "Response Three",
"Response Twelve", "Response Three", "Response Twelve", "Response Three",
"Response Twelve", "Response Three", "Response Twelve", "Response Three",
"Response Twelve", "Response Three", "Response Twelve", "Response Three",
"Response Twelve", "Response Three", "Response Twelve", "Response Three",
"Response Seven", "Response Seven", "Response Seven", "Response Eight",
"Response Three", "Response Two", "Response One", "Response Ten",
"Response Two", "Response Ten", "Response Two", "Response Ten",
"Response Two", "Response Three", "Response One", "Response Three",
"Response One", "Response Three", "Response One", "Response Eight",
"Response Nine", "Response Three", "Response Ten", "Response Two",
"Response One", "Response Eight", "Response Nine", "Response Three",
"Response Ten", "Response Two", "Response One", "Response Eight",
"Response Nine", "Response Three", "Response Ten", "Response Two",
"Response One", "Response Eight", "Response Nine", "Response Three",
"Response Ten", "Response Two", "Response One", "Response Eight",
"Response Nine", "Response Three", "Response Ten", "Response Two",
"Response One", "Response Eight", "Response Nine", "Response Three",
"Response Ten", "Response Two", "Response One", "Response Eight",
"Response Three", "Response Ten", "Response Eight", "Response Three",
"Response Ten", "Response Eight", "Response Three", "Response Ten",
"Response Eight", "Response Three", "Response Ten", "Response Eight",
"Response Nine", "Response Three", "Response Two", "Response Six",
"Response Eight", "Response Nine", "Response Three", "Response Two",
"Response Six", "Response Eight", "Response Nine", "Response Three",
"Response Two", "Response Six", "Response Twelve", "Response One",
"Response Twelve", "Response One", "Response Twelve", "Response One",
"Response Twelve", "Response One", "Response Seven", "Response Seven",
"Response Seven", "Response Seven", "Response Eight", "Response Nine",
"Response Twelve", "Response Eleven", "Response Ten", "Response Two",
"Response One", "Response Four", "Response Five", "Response Six",
"Response Eight", "Response Nine", "Response Ten", "Response One",
"Response Eight", "Response Nine", "Response Ten", "Response One",
"Response Eight", "Response Nine", "Response Ten", "Response One",
"Response Eight", "Response Nine", "Response Ten", "Response One",
"Response Eight", "Response Nine", "Response Ten", "Response One",
"Response Seven", "Response Seven", "Response Seven", "Response Seven",
"Response Eight", "Response Nine", "Response Twelve", "Response Ten",
"Response Eight", "Response Nine", "Response Twelve", "Response Ten",
"Response Eight", "Response Three", "Response Ten", "Response Eight",
"Response Three", "Response Ten", "Response Eight", "Response Three",
"Response Ten", "Response One", "Response One", "Response One",
"Response One", "Response Seven", "Response Eight", "Response Twelve",
"Response Ten", "Response Eight", "Response Twelve", "Response Ten",
"Response Eight", "Response Twelve", "Response Ten", "Response Eight",
"Response Twelve", "Response Ten", "Response Eight", "Response Twelve",
"Response Ten"), n = c(3210L, 4658L, 1271L, 4453L, 2834L, 2526L,
2526L, 3210L, 4098L, 4658L, 1271L, 4453L, 2975L, 2834L, 3833L,
916L, 1221L, 1208L, 3210L, 4098L, 4658L, 1271L, 4453L, 2975L,
2834L, 3833L, 916L, 1221L, 1208L, 3210L, 4098L, 4658L, 1271L,
4453L, 2975L, 2834L, 3833L, 916L, 1221L, 1208L, 3210L, 4098L,
4658L, 1271L, 4453L, 2975L, 2834L, 3833L, 916L, 1221L, 1208L,
2526L, 2526L, 4453L, 3833L, 4453L, 3833L, 4453L, 3833L, 4453L,
3833L, 4658L, 4453L, 4658L, 4453L, 4658L, 4453L, 4658L, 4453L,
4658L, 4453L, 4658L, 4453L, 4658L, 4453L, 4658L, 4453L, 4658L,
4453L, 4658L, 4453L, 4658L, 4453L, 4658L, 4453L, 2526L, 2526L,
2526L, 3210L, 4453L, 2834L, 3833L, 2975L, 2834L, 2975L, 2834L,
2975L, 2834L, 4453L, 3833L, 4453L, 3833L, 4453L, 3833L, 3210L,
4098L, 4453L, 2975L, 2834L, 3833L, 3210L, 4098L, 4453L, 2975L,
2834L, 3833L, 3210L, 4098L, 4453L, 2975L, 2834L, 3833L, 3210L,
4098L, 4453L, 2975L, 2834L, 3833L, 3210L, 4098L, 4453L, 2975L,
2834L, 3833L, 3210L, 4098L, 4453L, 2975L, 2834L, 3833L, 3210L,
4453L, 2975L, 3210L, 4453L, 2975L, 3210L, 4453L, 2975L, 3210L,
4453L, 2975L, 3210L, 4098L, 4453L, 2834L, 1208L, 3210L, 4098L,
4453L, 2834L, 1208L, 3210L, 4098L, 4453L, 2834L, 1208L, 4658L,
3833L, 4658L, 3833L, 4658L, 3833L, 4658L, 3833L, 2526L, 2526L,
2526L, 2526L, 3210L, 4098L, 4658L, 1271L, 2975L, 2834L, 3833L,
916L, 1221L, 1208L, 3210L, 4098L, 2975L, 3833L, 3210L, 4098L,
2975L, 3833L, 3210L, 4098L, 2975L, 3833L, 3210L, 4098L, 2975L,
3833L, 3210L, 4098L, 2975L, 3833L, 2526L, 2526L, 2526L, 2526L,
3210L, 4098L, 4658L, 2975L, 3210L, 4098L, 4658L, 2975L, 3210L,
4453L, 2975L, 3210L, 4453L, 2975L, 3210L, 4453L, 2975L, 3833L,
3833L, 3833L, 3833L, 2526L, 3210L, 4658L, 2975L, 3210L, 4658L,
2975L, 3210L, 4658L, 2975L, 3210L, 4658L, 2975L, 3210L, 4658L,
2975L)), row.names = c(NA, -250L), class = c("tbl_df", "tbl",
"data.frame"))
I then create a new data frame that is a bootstrapped confidence interval for median age of each value/response in the original data frame. I use the infer package to create the ci. Here is the code and the dput for the data frame.
bootstrapped_ci_df <- data_frame_responses %>%
split(.$value) %>%
map_df(~.x %>%
specify(response = Ages) %>%
generate(reps = 1000, type = 'bootstrap') %>%
calculate(stat = 'median') %>%
get_ci(level = 0.99999), .id = 'value')
structure(list(value = c("Response One", "Response Two", "Response Three",
"Response Four", "Response Five", "Response Six", "Response Seven",
"Response Eight", "Response Nine", "Response Ten", "Response Eleven",
"Response Twelve"), lower_ci = c(28.5, 28, 29, 28, 30, 29, 31,
29, 28, 29, 28, 28), upper_ci = c(29, 29, 30, 29, 30.995005,
30, 31.5, 30, 29, 30, 29, 29)), row.names = c(NA, -12L), class = c("tbl_df",
"tbl", "data.frame"))
The trouble is when I go to graph this with ggplot2.
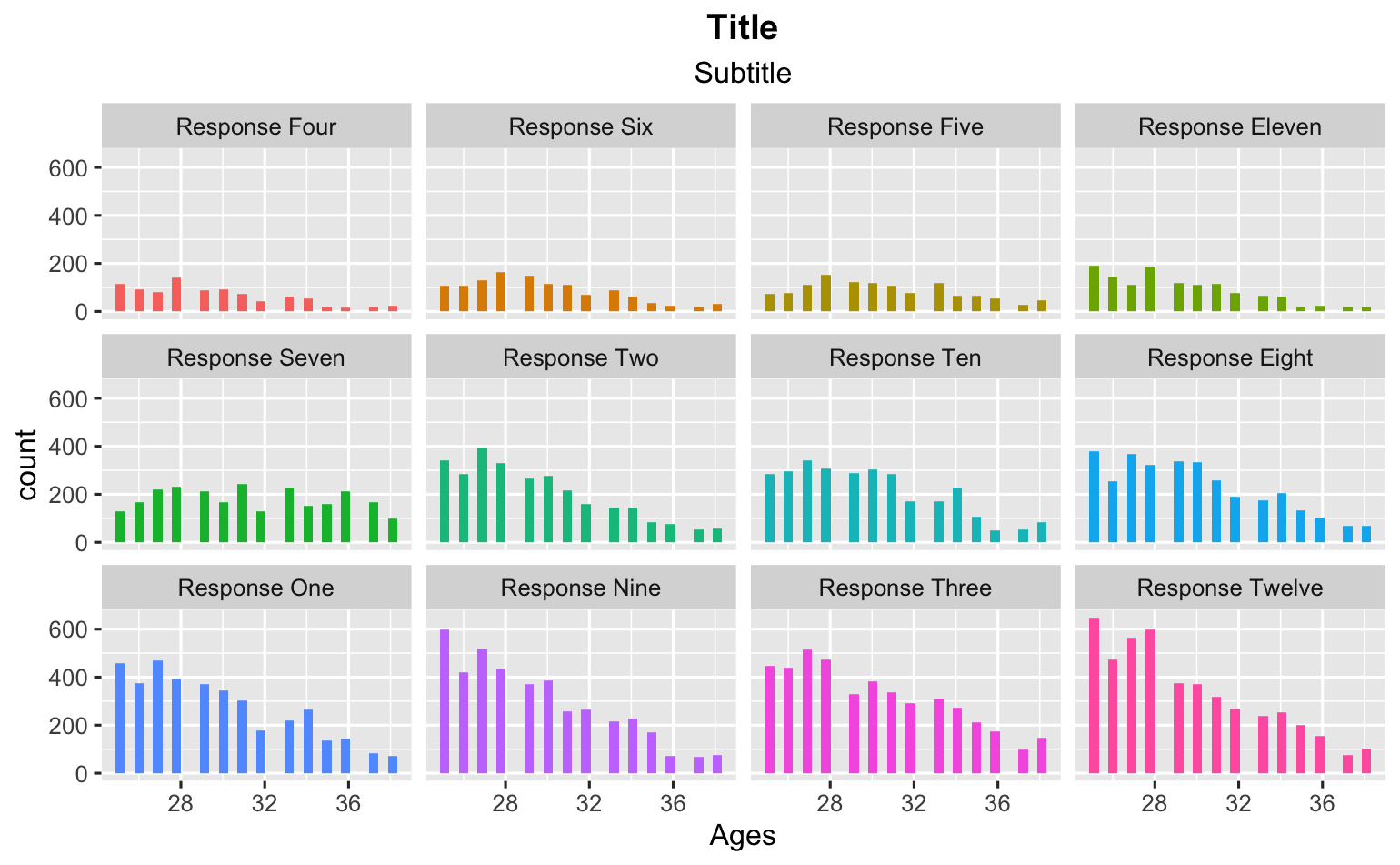
In the first case I can create a graph that looks good. I put the values with the most responses on the bottom.
ggplot(data_frame_responses, aes(x = Ages, fill = fct_reorder(value, n)))
geom_histogram()
facet_wrap(~ fct_reorder(value, n))
theme(legend.position="none")
labs(title = "Title")
labs(subtitle = "Subtitle")
theme(plot.title = element_text(hjust = 0.5, face = "bold"))
theme(plot.subtitle = element_text(hjust = 0.5))
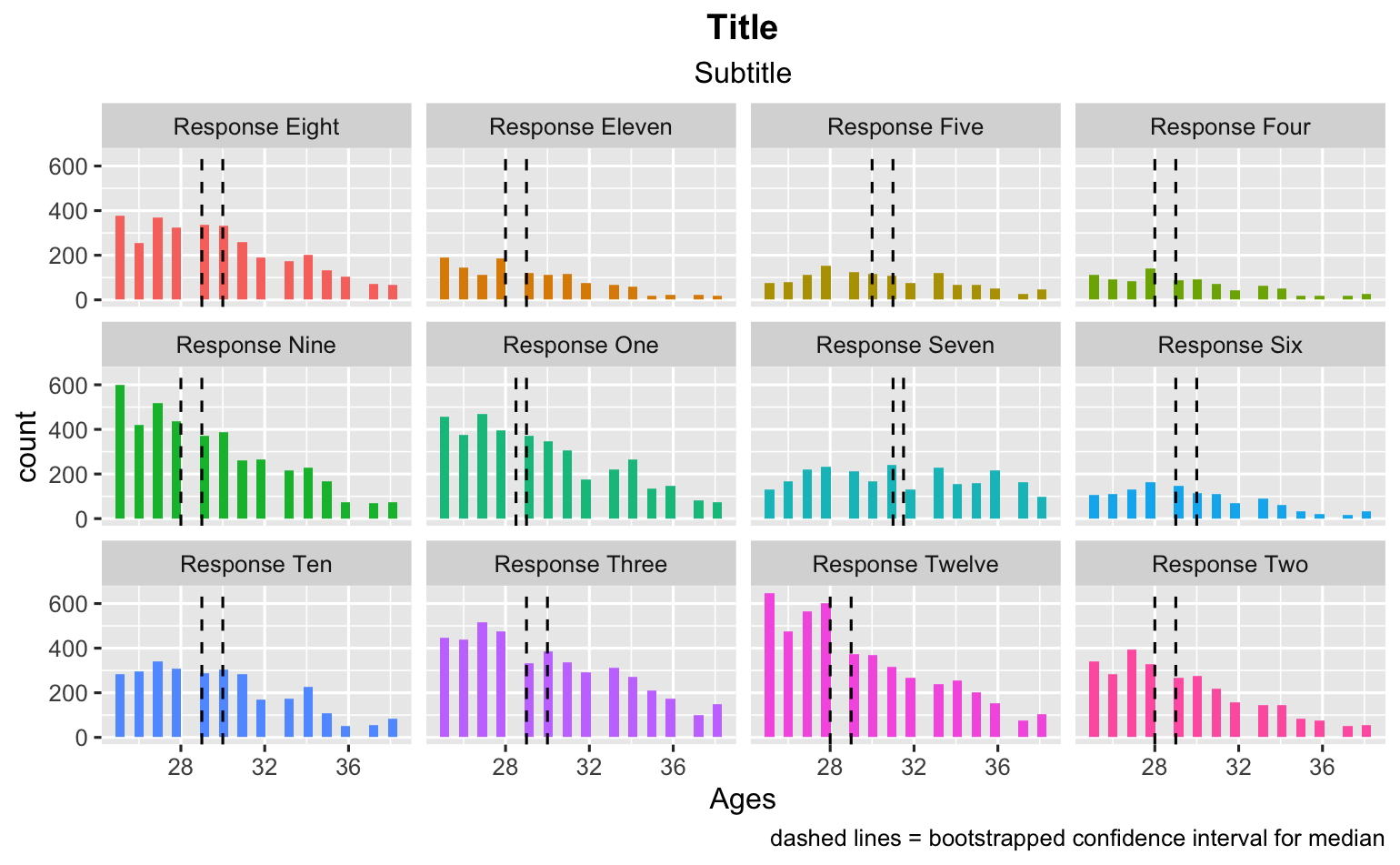
I can also map the confidence intervals on the data frame and that looks good, too.
ggplot(data_frame_responses, aes(x = Ages, fill = value))
geom_histogram()
facet_wrap(~value)
theme(legend.position="none")
labs(title = "Title")
labs(subtitle = "Subtitle")
geom_vline(mapping = aes(xintercept = lower_ci), bootstrapped_ci_df, linetype = 'dashed')
geom_vline(mapping = aes(xintercept = upper_ci), bootstrapped_ci_df, linetype = 'dashed')
labs(caption = "dashed lines = bootstrapped confidence interval for median")
theme(plot.title = element_text(hjust = 0.5, face = "bold"))
theme(plot.subtitle = element_text(hjust = 0.5))
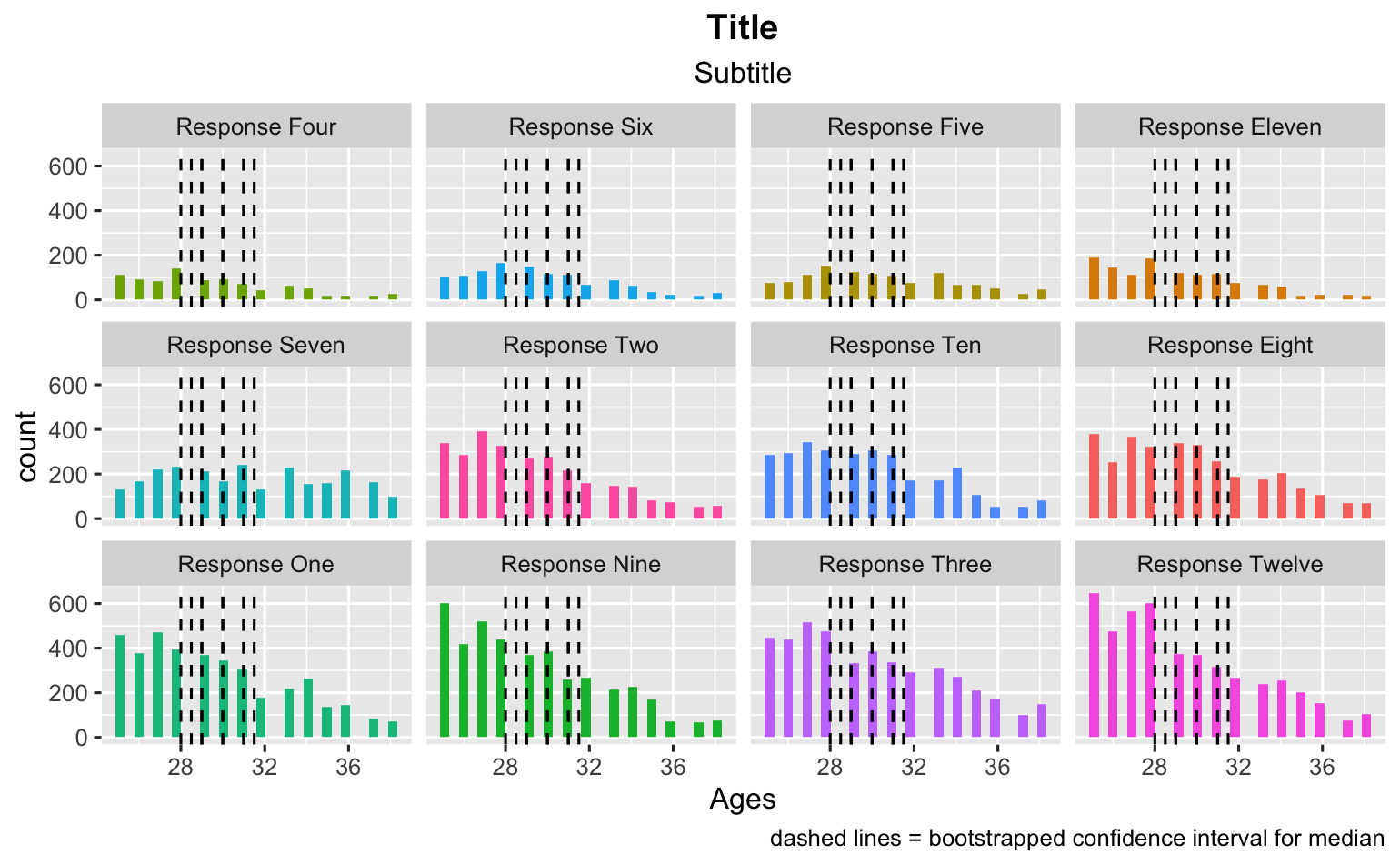
The trouble is, I can't figure out how to do both. How can I reorder the data frame with the facets and also keep the mapping in tact? Here is my try and you can see how it went wrong.
ggplot(data_frame_responses, aes(x = Ages, fill = value))
geom_histogram()
facet_wrap(~ fct_reorder(value, n))
theme(legend.position="none")
labs(title = "Title")
labs(subtitle = "Subtitle")
geom_vline(mapping = aes(xintercept = lower_ci), bootstrapped_ci_df, linetype = 'dashed')
geom_vline(mapping = aes(xintercept = upper_ci), bootstrapped_ci_df, linetype = 'dashed')
labs(caption = "dashed lines = bootstrapped confidence interval for median")
theme(plot.title = element_text(hjust = 0.5, face = "bold"))
theme(plot.subtitle = element_text(hjust = 0.5))
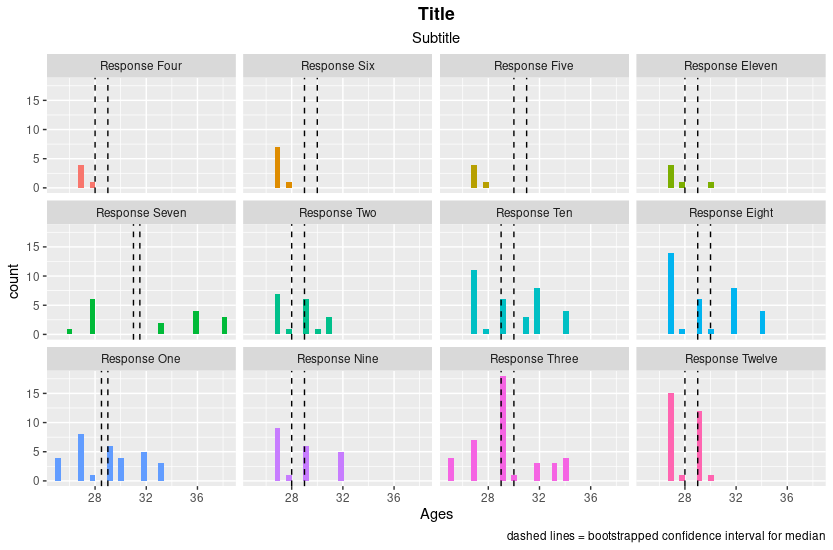
CodePudding user response:
Value variable in two data frames need to be consistent. In your code, you changed one data_frame_responses for geom_histogram, but not that for geom_vline
It is easier to change data types for both data.frames before ggplot. Please note: I used different data just for the purpose of answering your question.
data_frame_responses <-data_frame_responses %>% mutate(
value = fct_reorder(value, n)
)
bootstrapped_ci_df <-bootstrapped_ci_df %>%
mutate(value = factor(value, levels(data_frame_responses$value)))
ggplot(data_frame_responses, aes(x = Ages, fill = value))
geom_histogram()
facet_wrap(~value)
theme(legend.position="none")
labs(title = "Title")
labs(subtitle = "Subtitle")
geom_vline(mapping = aes(xintercept = lower_ci), bootstrapped_ci_df, linetype = 'dashed')
geom_vline(mapping = aes(xintercept = upper_ci), bootstrapped_ci_df, linetype = 'dashed')
labs(caption = "dashed lines = bootstrapped confidence interval for median")
theme(plot.title = element_text(hjust = 0.5, face = "bold"))
theme(plot.subtitle = element_text(hjust = 0.5))