I have the following dataset:
var1 = as.data.frame(c(runif(100,2,10),rep(NA,900)))
var2 = as.data.frame(runif(1000,-1,9))
colnames(var1)<-"var1"
colnames(var2)<-"var2"
data <- cbind(var1 ,var2)
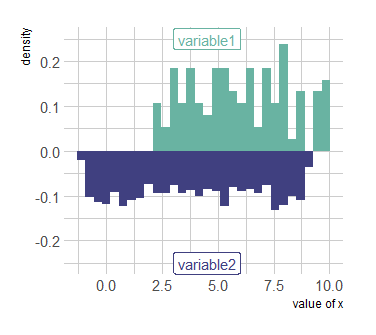
I want to plot the histogram of var1 and var2 in one plot, and as a mirror chart, without deleting rows from var2.
I used this code:
p <- ggplot(data, aes(x=x) )
geom_histogram( aes(x = var1, y = ..density..), fill="#69b3a2" )
geom_label( aes(x=4.5, y=0.25, label="variable1"), color="#69b3a2")
geom_histogram( aes(x = var2, y = -..density..), fill= "#404080")
geom_label( aes(x=4.5, y=-0.25, label="variable2"), color="#404080")
theme_ipsum()
xlab("value of x")
p
and I got this chart:
but it seems that this graph doesn't include 900 values of var2 (they were deleted because we have 900 NAs in var1).
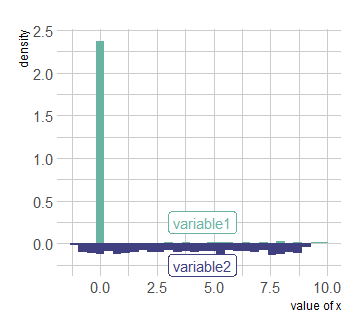
I don't want to replace the NAs with another value, because I will not have the required shape of graph, for example I replaced the NAs with 0 and this is what I got:
data[is.na(data)]<-0
Is there any way to plot the graph with all values in the dataset, and get the required plot which should be similar to the first plot ?
CodePudding user response:
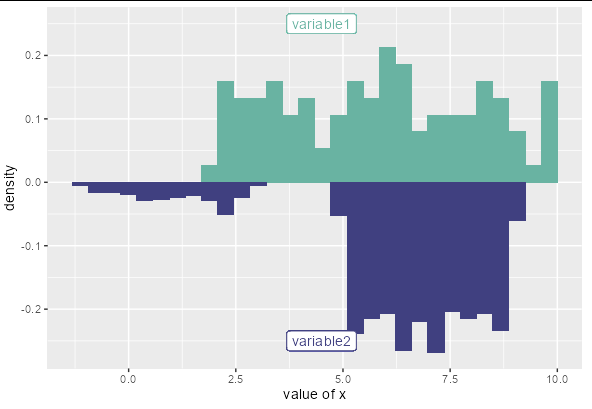
I think you are just getting confused with the warning message. The var2 data is being plotted. To reassure you of this, let's modify your data frame:
var1 = as.data.frame(c(runif(100,2,10), rep(NA, 900)))
var2 = as.data.frame(c(runif(100, -1, 3), runif(900, 5, 9)))
Now you can see that if all the rows where var1 is NA are removed, you should only see var2 values between -1 and 3. If var2 is plotted even though var1 is NA, we should also get some values between 5 and 9 being plotted for var2:
colnames(var1)<-"var1"
colnames(var2)<-"var2"
data <- cbind(var1 ,var2)
p <- ggplot(data, aes(x=x) )
geom_histogram( aes(x = var1, y = ..density..), fill="#69b3a2" )
geom_label( aes(x=4.5, y=0.25, label="variable1"), color="#69b3a2")
geom_histogram( aes(x = var2, y = -..density..), fill= "#404080")
geom_label( aes(x=4.5, y=-0.25, label="variable2"), color="#404080")
xlab("value of x")
p
So you don't need to worry; only the variable with missing entries will not be plotted.