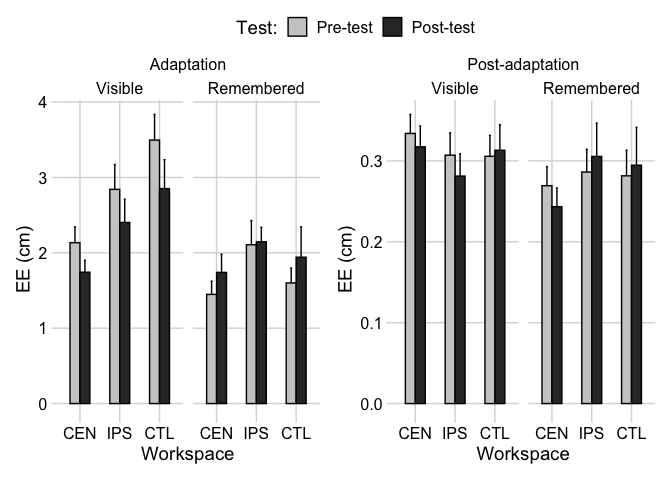
I am trying to make scales for the y-axis free. But it looks like it is not working. I want adaptation to have its own scale and post adaptation also having its own scales. I am trying to make scales for the y-axis free. But it looks like it is not working. I want adaptation to have its own scale and post adaptation also having its own scales. 
tgc <- structure(list(Group = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L), .Label = c("Visible", "Remembered"), class = "factor"),
Condition = structure(c(1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 3L,
3L, 3L, 3L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L
), .Label = c("CEN", "IPS", "CTL"), class = "factor"), test = structure(c(1L,
1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L,
1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L), .Label = c("Pre-test", "Post-test"
), class = "factor"), Session = structure(c(1L, 2L, 1L, 2L,
1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L,
2L, 1L, 2L, 1L, 2L), .Label = c("Adaptation", "Post-adaptation"
), class = "factor"), N = c(12, 12, 12, 12, 12, 12, 12, 12,
12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12,
12), EE = c(2.134379625, 0.333942625, 1.742841125, 0.317361916666667,
2.84197270833333, 0.307057416666667, 2.403112375, 0.281202,
3.49590529166667, 0.305657666666667, 2.85211466666667, 0.3131155,
1.44857545833333, 0.269328166666667, 1.740270875, 0.243361833333333,
2.10702266666667, 0.286209125, 2.145855125, 0.305474083333333,
1.60016616666667, 0.281528625, 1.94182179166667, 0.294655916666667
), sd = c(0.727246182828044, 0.0816168443914292, 0.549168068103643,
0.0894916121701392, 1.14554677132408, 0.0958562360654162,
1.06827971273128, 0.0953131237162305, 1.18204258551111, 0.0896670491921828,
1.32864473484909, 0.109865886496798, 0.605344957514288, 0.0815454655757737,
0.833908172662699, 0.0798994165789182, 1.11582277105041,
0.0976064300150272, 0.667812406644538, 0.142929179817685,
0.686043669971901, 0.109794818975944, 1.39509308576833, 0.161854932615856
), se = c(0.209937889711449, 0.0235607535398997, 0.158531165974993,
0.0258340031883217, 0.330690868396632, 0.0276713118479362,
0.308385789857611, 0.0275145288174349, 0.341226302469221,
0.0258846474942731, 0.383546697661249, 0.0317155495718416,
0.174748037086728, 0.0235401482506832, 0.240728553983119,
0.0230649748349663, 0.322110288616933, 0.0281765493219072,
0.192780836372198, 0.0412601002213964, 0.198043748767058,
0.0316950341456936, 0.402728684306467, 0.0467234944577166
), ci = c(0.462070179795855, 0.0518568689018959, 0.348924743722983,
0.0568602576432562, 0.727845693918804, 0.0609041467375754,
0.678752547059741, 0.0605590696140879, 0.751034027967696,
0.0569717250090983, 0.844180589754564, 0.069805453951774,
0.384617836383033, 0.0518115169661108, 0.529839974927164,
0.0507656673296478, 0.708959965158704, 0.0620161669201078,
0.424307760005262, 0.0908128682911871, 0.435891352085212,
0.0697602998032695, 0.886399857701764, 0.102837717929058)), row.names = c(NA,
-24L), class = "data.frame")
library(ggh4x)
p <- ggplot(tgc, aes(x = Condition, y = EE), fill = test)
geom_errorbar(aes(ymin=EE-se, ymax=EE se, group = test), position = position_dodge(0.5), width=.1)
geom_bar(aes(fill = test), stat = "identity", width = 0.5, color = "black", position='dodge') ylim(0,4) theme_bw() theme(
axis.text.x = element_text(size = 12,face="bold"),#, angle = 10, hjust = .5, vjust = .5),
axis.text.y = element_text(size = 12, face = "bold"),
axis.title.y = element_text(vjust= 1.8, size = 16),
axis.title.x = element_text(vjust= -0.5, size = 16),
axis.title = element_text(face = "bold")) xlab("Workspace") ylab("EE (cm)") theme(legend.position="top")
scale_fill_manual(values = c("grey80", "grey20")) facet_nested(. ~ Session Group, scales = "free_y") theme(aspect.ratio = 6/4)
p guides(fill=guide_legend(title="Test:")) theme(legend.text=element_text(size=14),legend.title=element_text(size=14) )
theme(strip.text = element_text(face="bold", size=12))
CodePudding user response:
The facet_nested() function is based on facet_grid(), in which you can have y scales that vary between rows in the grid, but not within a row.
Let's make the base of the plot.
library(ggplot2)
#> Warning: package 'ggplot2' was built under R version 4.1.1
library(ggh4x)
tgc <- structure(list(Group = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L), .Label = c("Visible", "Remembered"), class = "factor"),
Condition = structure(c(1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 3L,
3L, 3L, 3L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L
), .Label = c("CEN", "IPS", "CTL"), class = "factor"), test = structure(c(1L,
1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L,
1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L), .Label = c("Pre-test", "Post-test"
), class = "factor"), Session = structure(c(1L, 2L, 1L, 2L,
1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L,
2L, 1L, 2L, 1L, 2L), .Label = c("Adaptation", "Post-adaptation"
), class = "factor"), N = c(12, 12, 12, 12, 12, 12, 12, 12,
12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12, 12,
12), EE = c(2.134379625, 0.333942625, 1.742841125, 0.317361916666667,
2.84197270833333, 0.307057416666667, 2.403112375, 0.281202,
3.49590529166667, 0.305657666666667, 2.85211466666667, 0.3131155,
1.44857545833333, 0.269328166666667, 1.740270875, 0.243361833333333,
2.10702266666667, 0.286209125, 2.145855125, 0.305474083333333,
1.60016616666667, 0.281528625, 1.94182179166667, 0.294655916666667
), sd = c(0.727246182828044, 0.0816168443914292, 0.549168068103643,
0.0894916121701392, 1.14554677132408, 0.0958562360654162,
1.06827971273128, 0.0953131237162305, 1.18204258551111, 0.0896670491921828,
1.32864473484909, 0.109865886496798, 0.605344957514288, 0.0815454655757737,
0.833908172662699, 0.0798994165789182, 1.11582277105041,
0.0976064300150272, 0.667812406644538, 0.142929179817685,
0.686043669971901, 0.109794818975944, 1.39509308576833, 0.161854932615856
), se = c(0.209937889711449, 0.0235607535398997, 0.158531165974993,
0.0258340031883217, 0.330690868396632, 0.0276713118479362,
0.308385789857611, 0.0275145288174349, 0.341226302469221,
0.0258846474942731, 0.383546697661249, 0.0317155495718416,
0.174748037086728, 0.0235401482506832, 0.240728553983119,
0.0230649748349663, 0.322110288616933, 0.0281765493219072,
0.192780836372198, 0.0412601002213964, 0.198043748767058,
0.0316950341456936, 0.402728684306467, 0.0467234944577166
), ci = c(0.462070179795855, 0.0518568689018959, 0.348924743722983,
0.0568602576432562, 0.727845693918804, 0.0609041467375754,
0.678752547059741, 0.0605590696140879, 0.751034027967696,
0.0569717250090983, 0.844180589754564, 0.069805453951774,
0.384617836383033, 0.0518115169661108, 0.529839974927164,
0.0507656673296478, 0.708959965158704, 0.0620161669201078,
0.424307760005262, 0.0908128682911871, 0.435891352085212,
0.0697602998032695, 0.886399857701764, 0.102837717929058)), row.names = c(NA,
-24L), class = "data.frame")
p <- ggplot(tgc, aes(x = Condition, y = EE), fill = test)
geom_errorbar(aes(ymin=EE-se, ymax=EE se, group = test),
position = position_dodge(0.5), width=.1)
geom_bar(aes(fill = test),
stat = "identity", width = 0.5, color = "black", position='dodge')
theme_bw() theme(
axis.text.x = element_text(size = 12,face="bold"),
axis.text.y = element_text(size = 12, face = "bold"),
axis.title.y = element_text(vjust= 1.8, size = 16),
axis.title.x = element_text(vjust= -0.5, size = 16),
axis.title = element_text(face = "bold"))
xlab("Workspace") ylab("EE (cm)")
theme(legend.position="top")
scale_fill_manual(values = c("grey80", "grey20"))
theme(aspect.ratio = 6/4)
guides(fill=guide_legend(title="Test:"))
theme(legend.text=element_text(size=14),legend.title=element_text(size=14) )
theme(strip.text = element_text(face="bold", size=12))
If you wanted to have a non-grid layout, it is probably easier to use facet_nested_wrap() instead.
p facet_nested_wrap(~Session Group, scales = "free_y", nrow = 1)
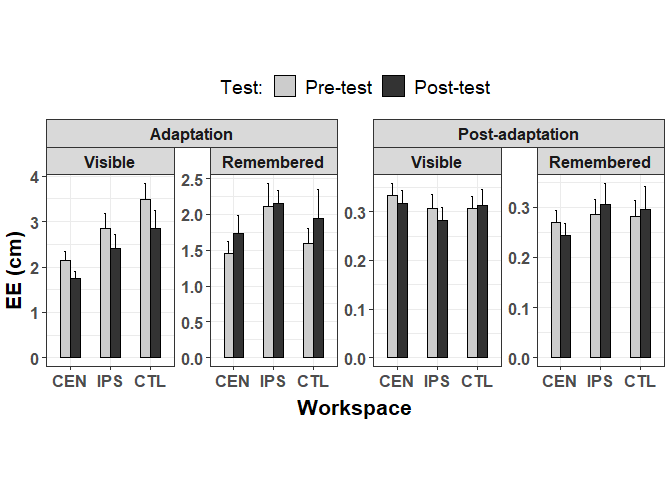
Alternatively, if you must retain the grid layout and want the y-axes to be independent within a row, you can use independent = "y".
p facet_nested(~Session Group, scales = "free_y", independent = "y")
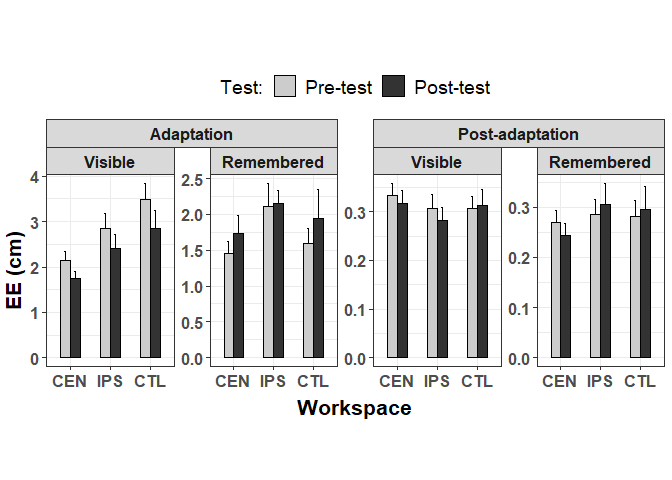
Created on 2021-11-09 by the reprex package (v2.0.1)
CodePudding user response:
I know it is not directly an answer to that question, but an alternative would be to create the panels separately. You can do this semi-programmatically by creating a list first. The biggest disadvantage that I can see in this way is that patchwork still cannot merge / combine x-/y- axis titles like it does with legends, so you would need to create each plot separately which would in turn defeat the point of this approach...
library(tidyverse)
library(ggh4x)
library(patchwork)
ls_p <-
tgc %>%
split(., .$Session) %>%
map(function(x){
## I've tried to de-clutter your graph. I am using cowplot::theme_minimal because it's a well thought through theme
ggplot(x, aes(x = Condition, y = EE), fill = test)
geom_errorbar(aes(ymin = EE - se, ymax = EE se, group = test), position = position_dodge(0.5), width = .1)
geom_bar(aes(fill = test), stat = "identity", width = 0.5, color = "black", position = "dodge")
labs(x= "Workspace", y = "EE (cm)")
scale_fill_manual("Test:", values = c("grey80", "grey20"))
facet_nested(. ~ Session Group, scales = "free_y")
cowplot::theme_minimal_grid()
})
wrap_plots(ls_p) plot_layout(guides = "collect") &
theme(legend.position = "top")