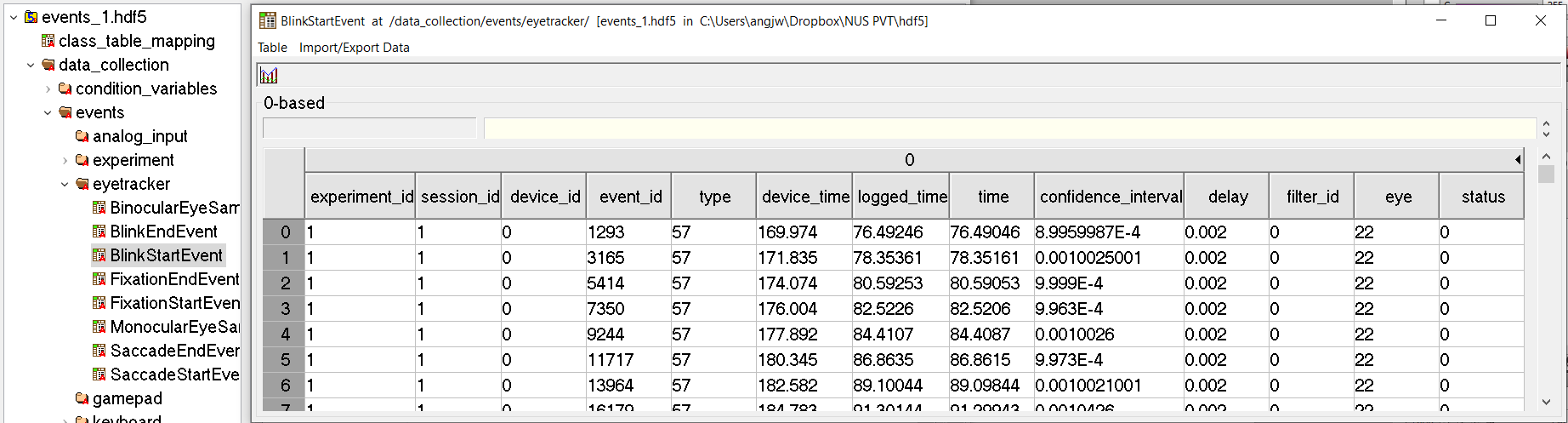
Below is a screenshot of the branches of data in my .hdf5 file. I am trying to extract the existing column names (ie. experiment_id, session_id....) from this particular BlinkStartEvent segment.
I have the following codes that was able to access to this section of the data and extract the numerical data as well. But for some reason, I cannot extract the corresponding column names, which I wish to append onto a separate list so I can create a dictionary out of this entire dataset. I thought .keys() was supposed to do it, but it didn't.
import h5py
def traverse_datasets(hdf_file):
def h5py_dataset_iterator(g, prefix=''):
for key in g.keys():
#print(key)
item = g[key]
path = f'{prefix}/{key}'
if isinstance(item, h5py.Dataset): # test for dataset
yield (path, item)
elif isinstance(item, h5py.Group): # test for group (go down)
yield from h5py_dataset_iterator(item, path)
for path, _ in h5py_dataset_iterator(hdf_file):
yield path
with h5py.File(filenameHDF[0], 'r') as f:
for dset in traverse_datasets(f):
if str(dset[-15:]) == 'BlinkStartEvent':
print('-----Path:', dset) # path that leads to the data
print('-----Shape:', f[dset].shape) #the length dimension of the data
print('-----Data type:', f[dset].dtype) #prints out the unicode for all columns
data2 = f[dset][()] # The entire dataset
# print('Check column names', f[dset].keys()) # I tried this but I got a AttributeError: 'Dataset' object has no attribute 'keys' error
I got the following as the output:
-----Path: /data_collection/events/eyetracker/BlinkStartEvent
-----Shape: (220,)
-----Data type: [('experiment_id', '<u4'), ('session_id', '<u4'), ('device_id', '<u2'), ('event_id', '<u4'), ('type', 'u1'), ('device_time', '<f4'), ('logged_time', '<f4'), ('time', '<f4'), ('confidence_interval', '<f4'), ('delay', '<f4'), ('filter_id', '<i2'), ('eye', 'u1'), ('status', 'u1')]
Traceback (most recent call last):
File "C:\Users\angjw\Dropbox\NUS PVT\Analysis\PVT analysis_hdf5access.py", line 64, in <module>
print('Check column names', f[dset].keys())
AttributeError: 'Dataset' object has no attribute 'keys'
What am I getting wrong here?
Also, is there a more efficient way to access the data such that I can do something (hypothetical) like:
data2[0]['experiment_id'] = 1
data2[1]['time'] = 78.35161
data2[2]['logged_time'] = 80.59253
rather than having to go through the process of setting up a dictionary for every single row of data?
CodePudding user response:
You're close. The dataset's .dtype gives you the dataset as a NumPy dtype. Adding .descr returns it as a list of (field name, field type) tuples. See code below to print the field names inside your loop:
for (f_name,f_type) in f[dset].dtype.descr:
print(f_name)
There are better ways to work with HDF5 data than creating a dictionary for every single row of data (unless you absolutely want a dictionary for some reason). h5py is designed to work with dataset objects similar to NumPy arrays. (However, not all NumPy operations work on h5py dataset objects). The following code accesses the data and returns 2 similar (but slightly different) data objects.
# this returns a h5py dataset object that behaves like a NumPy array:
dset_obj = f[dset]
# this returns a NumPy array:
dset_arr = f[dset][()]
You can slice data from either object using standard NumPy slicing notation (using field names and row values). Continuing from above...
# returns row 0 from field 'experiment_id'
val0 = dset_obj[0]['experiment_id']
# returns row 1 from field 'time'
val1 = dset_obj[1]['time']
# returns row 2 from field 'logged_time'
val2 = dset_obj[2]['logged_time']
(You will get the same values if you replace dset_obj with dset_arr above.)
You can also slice entire fields/columns like this:
# returns field 'experiment_id' as a NumPy array
expr_arr = dset_obj['experiment_id']
# returns field 'time' as a NumPy array
time_arr = dset_obj['time']
# returns field 'logged_time' as a NumPy array
logtime_arr = dset_obj['logged_time']
That should answer your initial questions. If not, please add comments (or modify the post), and I will update my answer.
CodePudding user response:
My previous answer used the h5py package (same package as your code). There is another Python package that I like to use with HDF5 data: PyTables (aka tables). Both are very similar, and each has unique strengths.
- h5py attempts to map the HDF5 feature set to NumPy as closely as possible. Also, it uses Python dictionary syntax to iterate over object names and values. So, it is easy to learn if you are familiar with NumPy. Otherwise, you have to learn some NumPy basics (like interrogating dtypes). Homogeneous data is returned as a
np.arrayand heterogeneous data (like yours) is returned as anp.recarray. - PyTables builds an additional abstraction layer on top of HDF5 and NumPy. Two unique capabilities I like are: 1) recursive iteration over nodes (groups or datasets), so a custom dataset generator isn't required, and 2) heterogeneous data is accessed with a "Table" object that has more methods than basic NumPy recarray methods. (Plus it can do complex queries on tables, has advanced indexing capabilities, and is fast!)
To compare them, I rewrote your h5py code with PyTables so you can "see" the difference. I incorporated all the operations in your question, and included the equivalent calls from my h5py answer. Differences to note:
- The
f.walk_nodes()method is a built-in method that replaces your your generator. However, it returns an object (a Table object in this case), not the Table (dataset) name. So, the code is slightly different to work with the object instead of the name. - Use
Table.read()to load the data into a NumPy (record) array. Different examples show how to load the entire Table into an array, or load a single column referencing the field name.
Code below:
import tables as tb
with tb.File(filenameHDF[0], 'r') as f:
for tb_obj in f.walk_nodes('/','Table'):
if str(tb_obj.name[-15:]) == 'BlinkStartEvent':
print('-----Name:', tb_obj.name) # Table name without the path
print('-----Path:', tb_obj._v_pathname) # path that leads to the data
print('-----Shape:', tb_obj.shape) # the length dimension of the data
print('-----Data type:', tb_obj.dtype) # prints out the np.dtype for all column names/variable types
print('-----Field/Column names:', tb_obj.colnames) #prints out the names of all columns as a list
data2 = tb_obj.read() # The entire Table (dataset) into array data2
# returns field 'experiment_id' as a NumPy (record) array
expr_arr = tb_obj.read(field='experiment_id')
# returns field 'time' as a NumPy (record) array
time_arr = tb_obj.read(field='time')
# returns field 'logged_time' as a NumPy (record) array
logtime_arr = tb_obj.read(field='logged_time')