The title may not be the full description of what I want to do, but I will try to fully explain what I am working with. I have a pandas Dataframe that contains all data from files in a directory.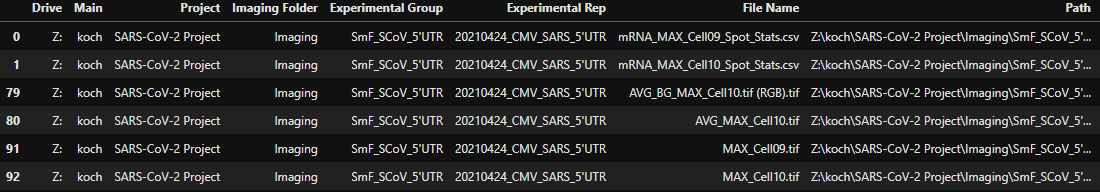
> Drive Main Project Imaging Folder Experimental Group \ > 0 Z: koch SARS-CoV-2 Project Imaging SmF_SCoV_5'UTR > 1 Z: koch SARS-CoV-2 Project Imaging SmF_SCoV_5'UTR > 79 Z: koch SARS-CoV-2 Project Imaging SmF_SCoV_5'UTR > 80 Z: koch SARS-CoV-2 Project Imaging SmF_SCoV_5'UTR > 91 Z: koch SARS-CoV-2 Project Imaging SmF_SCoV_5'UTR > 92 Z: koch SARS-CoV-2 Project Imaging SmF_SCoV_5'UTR > > > Experimental Rep File Name \ 0 20210424_CMV_SARS_5'UTR mRNA_MAX_Cell09_Spot_Stats.csv 1 > 20210424_CMV_SARS_5'UTR mRNA_MAX_Cell10_Spot_Stats.csv 79 > 20210424_CMV_SARS_5'UTR AVG_BG_MAX_Cell10.tif (RGB).tif 80 > 20210424_CMV_SARS_5'UTR AVG_MAX_Cell10.tif 91 > 20210424_CMV_SARS_5'UTR MAX_Cell09.tif 92 > 20210424_CMV_SARS_5'UTR MAX_Cell10.tif > > Path 7 0 Z:\koch\SARS-CoV-2 Project\Imaging\SmF_SCoV_5'... NaN 1 > Z:\koch\SARS-CoV-2 Project\Imaging\SmF_SCoV_5'... NaN 79 > Z:\koch\SARS-CoV-2 Project\Imaging\SmF_SCoV_5'... None 80 > Z:\koch\SARS-CoV-2 Project\Imaging\SmF_SCoV_5'... None 91 > Z:\koch\SARS-CoV-2 Project\Imaging\SmF_SCoV_5'... None 92 > Z:\koch\SARS-CoV-2 Project\Imaging\SmF_SCoV_5'... None
In this dataframe I have both csv files and tif files. I already have code that will read in all the csv files and I can access them based on file file name.
FINDPATH = Path("Z:\koch\Imaging")
FILEEXT = ("*.csv")
files_dfs = {}
for csv_file in FINDPATH.rglob(FILEEXT):
filename = csv_file.name
df = pd.read_csv(csv_file)
files_dfs[filename] = df
Each of these csv files have a respective tif file. For instance mRNA_MAX_Cell09_Spot_Stats.csv comes from MAX_Cell09.tif.
I now need to come up with a way to iterate over the dataframe, take only the original tif file (all named MAX_Cell...), output the filepath, and finally get the respective csv File Name.
The goal is to be able to take the tif file and do in silico simulations and compare to the real data in the csv file.
I hope this is well explained. Any insight would be more than helpful.
Thank you
CodePudding user response:
There are basically two steps here:
- Get the cell number so we can match files belonging to the same experiment and cell number.
- Use groupby() to perform the matching.
After that, you can loop over the groupby, and get rows belonging to a single experiment.
Example:
s = """Drive,Main,Project,Imaging Folder,Experimental Group,Experimental Rep,File Name,Path
0,Z:,koch,SARS-CoV-2 Project,Imaging,SmF_SCoV_5'UTR,20210424_CMV_SARS_5'UTR,mRNA_MAX_Cell09_Spot_Stats.csv,Z:\koch\SARS-CoV-2 Project\Imaging\SmF_SCoV_5'UTR\mRNA_MAX_Cell09_Spot_Stats.csv
1,Z:,koch,SARS-CoV-2 Project,Imaging,SmF_SCoV_5'UTR,20210424_CMV_SARS_5'UTR,mRNA_MAX_Cell10_Spot_Stats.csv,Z:\koch\SARS-CoV-2 Project\Imaging\SmF_SCoV_5'UTR\mRNA_MAX_Cell10_Spot_Stats.csv
79,Z:,koch,SARS-CoV-2 Project,Imaging,SmF_SCoV_5'UTR,20210424_CMV_SARS_5'UTR,AVG_BG_MAX_Cell10.tif (RGB).tif,Z:\koch\SARS-CoV-2 Project\Imaging\SmF_SCoV_5'UTR\AVG_BG_MAX_Cell10.tif (RGB).tif
80,Z:,koch,SARS-CoV-2 Project,Imaging,SmF_SCoV_5'UTR,20210424_CMV_SARS_5'UTR,AVG_MAX_Cell10.tif,Z:\koch\SARS-CoV-2 Project\Imaging\SmF_SCoV_5'UTR\AVG_MAX_Cell10.tif
91,Z:,koch,SARS-CoV-2 Project,Imaging,SmF_SCoV_5'UTR,20210424_CMV_SARS_5'UTR,MAX_Cell09.tif,Z:\koch\SARS-CoV-2 Project\Imaging\SmF_SCoV_5'UTR\MAX_Cell09.tif
92,Z:,koch,SARS-CoV-2 Project,Imaging,SmF_SCoV_5'UTR,20210424_CMV_SARS_5'UTR,MAX_Cell10.tif,Z:\koch\SARS-CoV-2 Project\Imaging\SmF_SCoV_5'UTR\MAX_Cell10.tif
"""
import pandas as pd
import io
df = pd.read_csv(io.StringIO(s))
# Extract the cell number in the filename of each file
df['Cell Num'] = df['File Name'].str.extract('Cell([0-9] )[._]')
# Extract the filetype for each file
df['File Type'] = df['File Name'].str.extract('\\.([a-z]{2,4})')
# Keep only files which are either csv or tif files of the form MAX_CellNN.tif
df = df.loc[(df['File Type'] == 'csv') | (df['File Name'].str.match("MAX_Cell[0-9] .tif"))]
for group_id, group_df in df.groupby(["Experimental Group", "Experimental Rep", "Cell Num"]):
experimental_group, experimental_rep, cell_num = group_id
print(experimental_group, experimental_rep, cell_num)
file_group = group_df['File Name'].to_list()
print(file_group)
(The first eight lines are just to make the example reproducible. You don't need them if you already have the dataframe.)
