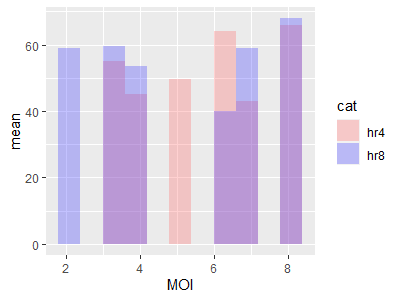
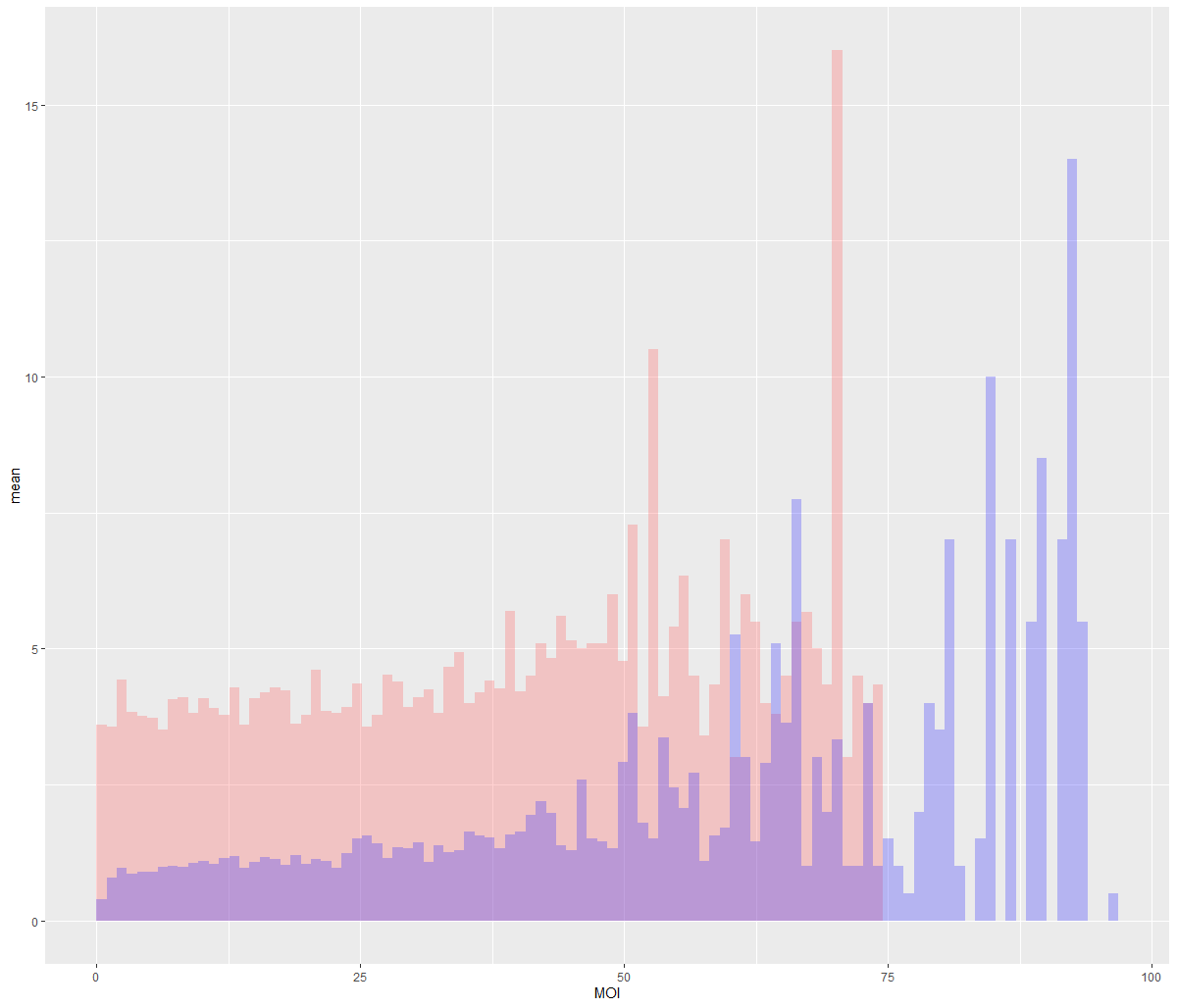
I came up with the following script to bin my data on X values, and plot the means of those bins in overlapping bar graphs. It works fine, but I can't seem to get a legend to generate, probably due to poor understanding of aesthetic mapping.
Here is the script, note that "MOI" and "T_cell_contacts" are two data columns in each DF.
ggplot(mapping=aes(MOI, T_cell_contacts)) stat_summary_bin(data = Cleaned24hr4, fun = "mean", geom="bar", bins= 100, fill = "#FF6666", alpha = 0.3) stat_summary_bin(data = cleaned24hr8, fun = "mean", geom="bar", bins= 100, fill = "#3733FF", alpha = 0.3) ylab("mean")
I also added the graph that it plots.
CodePudding user response:
I think the difficulty has to do with building a single legend out of two different geoms. My approach was to combine your data into a single data frame. The records from each to be set apart by a new category column, I'll call "cat" for short. With the popular dplyr package:
Cleaned24hr4 <- mutate(Cleaned24hr4, cat = "hr4")
Cleaned24hr8 <- mutate(Cleaned24hr8, cat = "hr8")
Then put them together:
Cleaned <- union(Cleaned24hr4,Cleaned24hr8)
Define your colors:
colorcode <- c("hr4" = "#FF6666", "hr8" = "#3733FF")
Here's my ggplot statement:
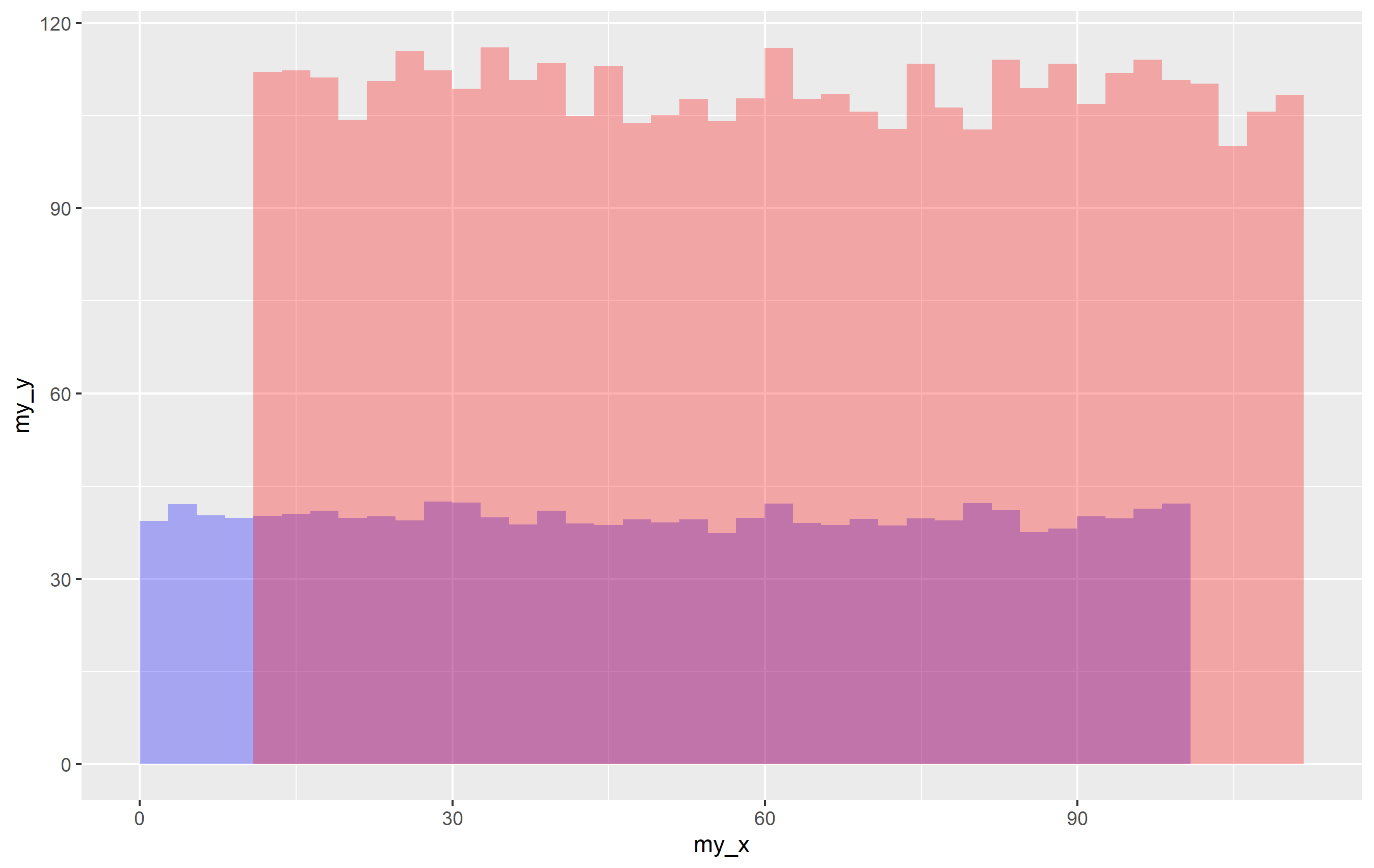
ggplot(Cleaned, mapping=aes(MOI, T_cell_contacts))
stat_summary_bin(fun = "mean", geom="bar", bins= 100, aes(fill = cat), alpha = 0.3)
scale_fill_manual(values = colorcode)
ylab("mean")
Output using some dummy data.
CodePudding user response:
Full disclosure: I was in the middle of writing this when @schumacher posted their response :). Decided to finish anyway.
There are two ways to approach this. One way (more complicated) is to keep the dataframes separate and ask ggplot2 to create a legend via mapping, and the second (simpler) way is to combine into one dataset similar to what @schumacher posted and map the fill color to the extra id column created.
I'll show you both, but first, here's a sample dataset:
library(ggplot2)
set.seed(8675309)
df1 <- data.frame(my_x=rep(1:100, 3), my_y=rnorm(300, 40, 4))
df2 <- data.frame(my_x=rep(11:110, 3), my_y=rnorm(300, 110, 10))
# and the plot code similar to OP's question
ggplot(mapping=aes(x = my_x, y = my_y))
stat_summary_bin(data=df1, fun="mean", geom="bar", bins=40, fill="blue", alpha=0.3)
stat_summary_bin(data=df2, fun="mean", geom="bar", bins=40, fill="red", alpha=0.3)
Method 1 : Combine Dataframes
This is the preferred method for a variety of reasons I can't list completely here. There are a lot of options you can use for combining datasets. One is using union() or rbind() after adding some sort of ID column to your data, but you can do all in one shot using bind_rows() from dplyr:
df <- dplyr::bind_rows(list(dataset1 = df1, dataset2 = df2), .id="id")
The result will bind the rows together and by specifying the .id argument, it will create a new column in the dataset called "id" that uses the names for each of the datasets in the list as the value. In this case, the value in thd df$id column is either "dataset1" if it originated from df1 or "dataset2" if it originated from df2.
Then you use aes(fill=...) to map the fill color to the column "id" in the combined dataset.
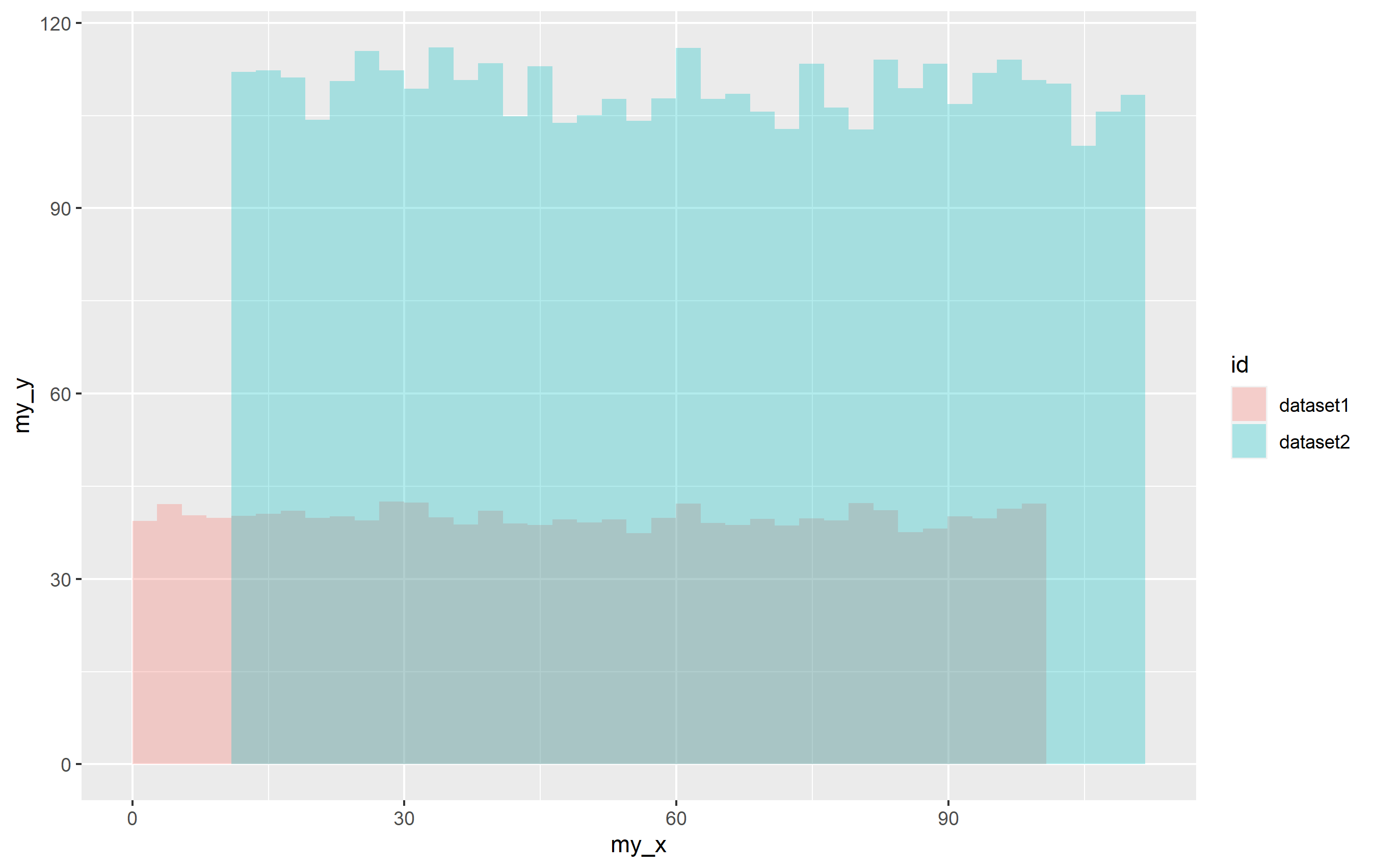
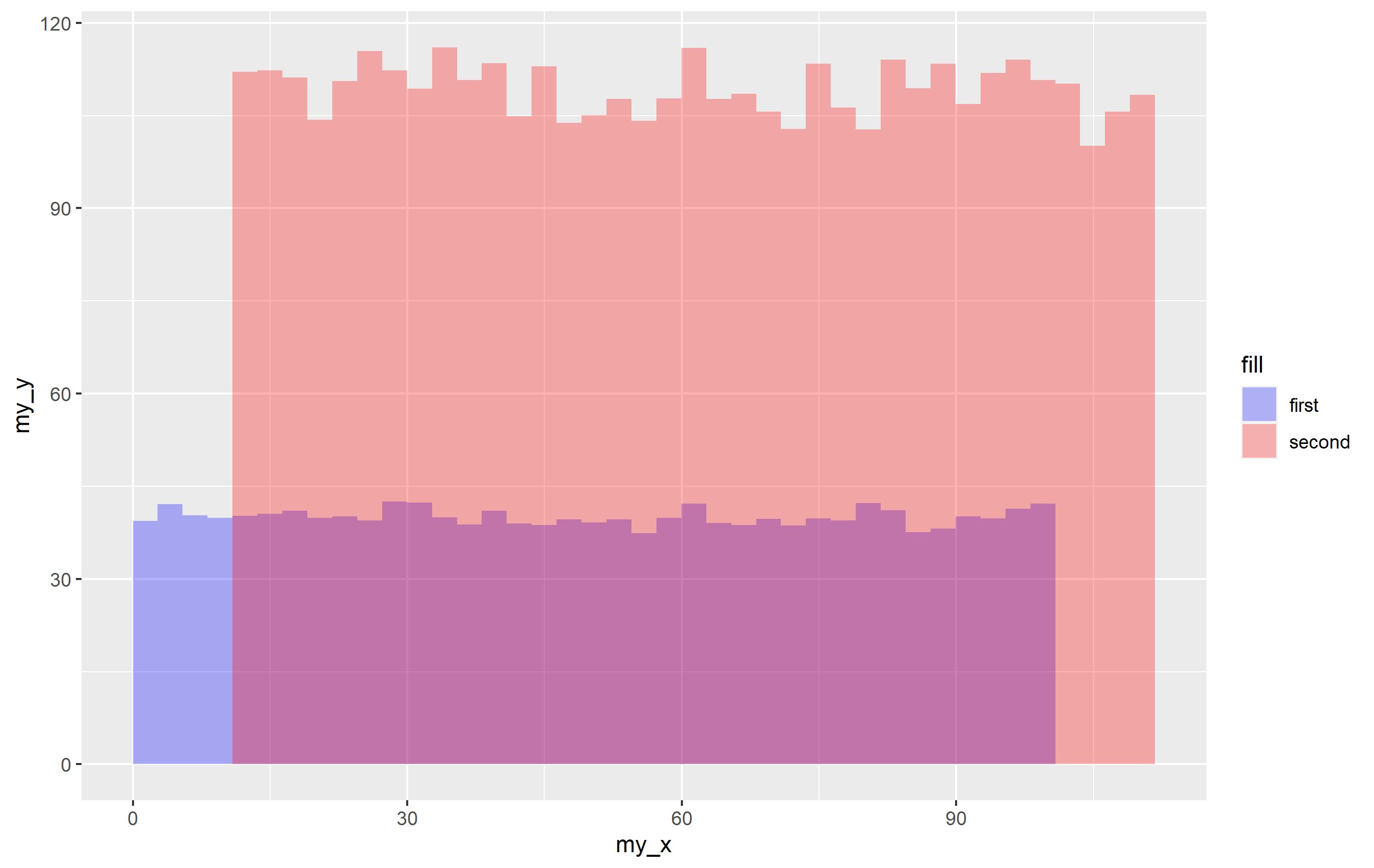
p <- ggplot(df, aes(x=my_x, y=my_y))
stat_summary_bin(aes(fill=id), fun="mean", geom="bar", bins=40, alpha=0.3)
p
This creates a plot with the default colors for fill, so if you want to supply your own, just use scale_fill_manual(values=...) to specify the particular colors. Using a named vector for values= ensures that each color is applied the way you want it to be, but you can just supply an unnamed vector of color names.
p scale_fill_manual(values = c("dataset1" = "blue", "dataset2" = "red"))
Method 2 : Use mapping to add the legend
While Method 1 is preferred, there is another way that does not force you to combine your dataframes. This is also useful to illustrate a bit about how ggplot2 decides to create and draw legends. The legend is created automaticaly via the mapping= argument, specifically via aes(). If you put any aesthetic inside of aes() that would normally impart a different appearance and not location (with some exceptions like x, y, and label), then this initiates the creation of a legend. You can map either a column in your dataset (like above), or you can just supply a single value and that will be applied to the entire dataset used for the geom. In this case, see what happens when you change the fill= argument for each geom call to be within aes() and assign it to a character value:
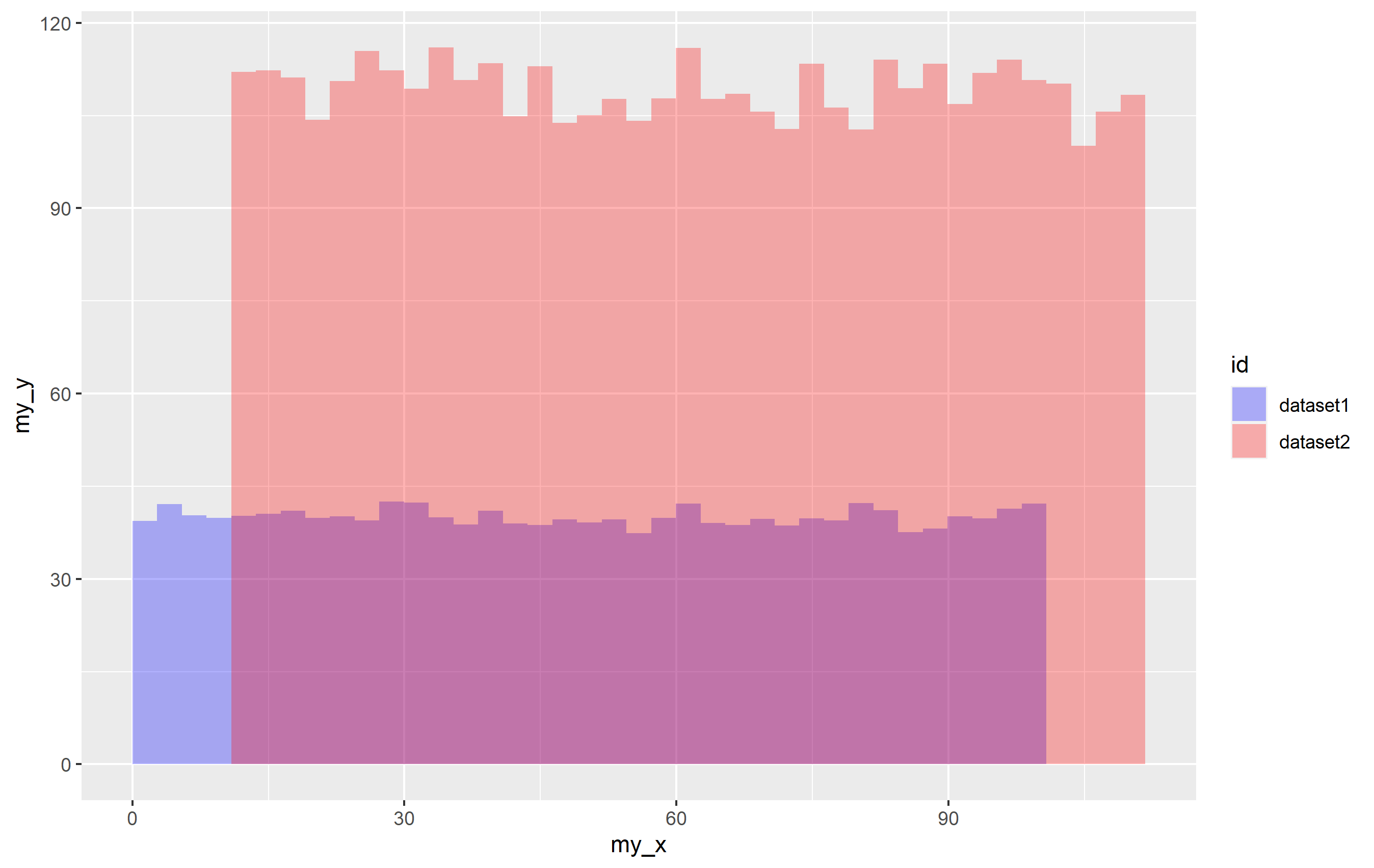
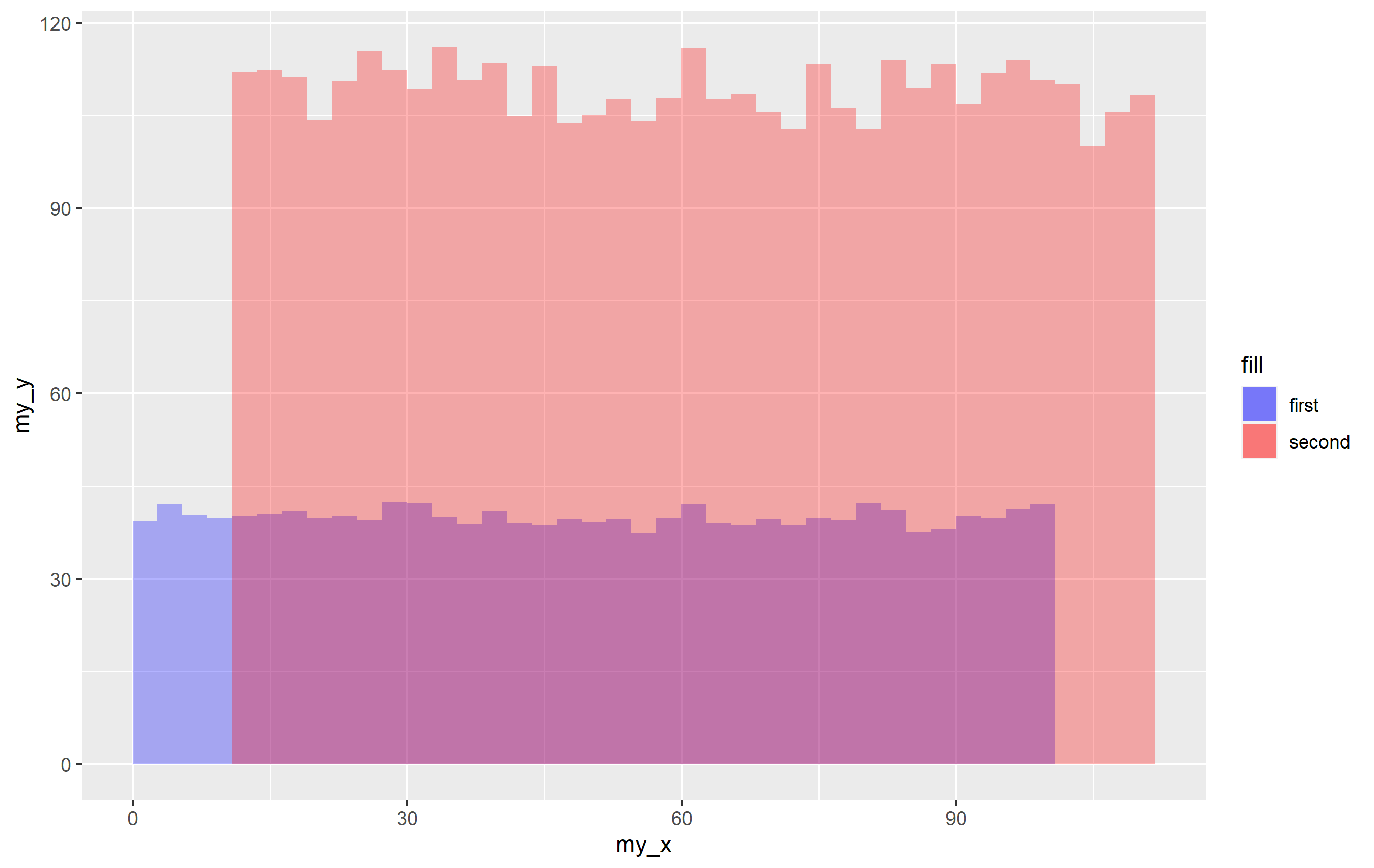
p1 <- ggplot(mapping = aes(x=my_x, y=my_y))
stat_summary_bin(aes(fill="first"), data=df1, fun="mean", geom="bar", bins=40, alpha=0.3)
stat_summary_bin(aes(fill="second"), data=df2, fun="mean", geom="bar", bins=40, alpha=0.3)
scale_fill_manual(values = c("first" = "blue", "second" = "red"))
p1
It works! When you provide a character value for the fill= aesthetic inside aes(), it's basically labeling every observation in that data to have the value "first" or "second" and using that to make the legend. Cool, right?
You notice a problem though, which is that the alpha value for the legend is not correct. This is because you get overplotting. It's just one of the reasons why you shouldn't really do it this way, but... sort of works. It is only noticeable if you ahve an alpha value. You can get that to look normal, but you need to use guide_legend() to override the aesthetics. Since the code effectively causes the legend to be drawn completely for each geom... you have to cut the alpha value in half for it to display correctly.
p1 guides(fill=guide_legend(override.aes = list(alpha=0.15)))
Oh, and the real reason why not to use Method 2 is.... just think about doing that again for 5 datasets... how about 10?... how about 20?.....