I'm currently trying to run tbl_regression with an imputed dataset from mice run through a logistic glm. I'm having trouble trying to combine the custom tidiers pool_and_tidy_mice and tidy_standardize in order to get the regression output from the pooled mice results and the standardized odds ratio estimates.
Is there a way to get the standardized odds ratios with pooling imputed data with tbl_regression or possibly another step I can take to get them?
Using the surival package as an example, I can get standardized odds ratio with non-imputed data with this code:
library(tidyverse)
library(broom.mixed)
library(broom)
library(survival)
mod <- glm(death ~ marker grade, data = trial, family = "binomial")
tbl_regression(mod, tidy_fun = tidy_standardize,
exponentiate = TRUE, conf.int = TRUE, conf.level = 0.95)
But, if I try to use tidy_standardize in the following code,
suppressWarnings(mice::mice(trial, m = 2)) %>%
with(glm(death ~ marker grade, family = "binomial")) %>%
tbl_regression(tidy_fun = tidy_standardize,
exponentiate = TRUE, conf.int = TRUE, conf.level = 0.95)
I get this error:
x There was an error calling `tidy_fun()`. Most likely, this is because the
function supplied in `tidy_fun=` was misspelled, does not exist, is not
compatible with your object, or was missing necessary arguments (e.g. `conf.level=` or `conf.int=`). See error message below.
Error: Error in .model_parameters_generic(model = mice::pool(model), ci = ci, : formal argument "standardize" matched by multiple actual arguments
In addition: Warning messages:
1: Could not get model data.
2: No variables could be standardized.
3: Could not get model data.
4: No variables could be standardized.
I also tried the following but still got an error message:
suppressWarnings(mice::mice(trial, m = 2)) %>%
with(glm(death ~ marker grade, family = "binomial")) %>%
tbl_regression(tidy_fun = purrr::partial(tidy_standardize, method = "posthoc"),
exponentiate = TRUE, conf.int = TRUE, conf.level = 0.95)
x There was an error calling `tidy_fun()`. Most likely, this is because the
function supplied in `tidy_fun=` was misspelled, does not exist, is not
compatible with your object, or was missing necessary arguments (e.g. `conf.level=` or `conf.int=`). See error message below.
Error: Error in .model_parameters_generic(model = mice::pool(model), ci = ci, : formal argument "standardize" matched by multiple actual arguments
CodePudding user response:
Unlike the unimputed data, mice doesn't output a table, so it must be transformed using complete afterwards. What about this:
library(tidyverse)
library(broom.mixed)
library(broom)
library(survival)
library(gtsummary)
library(mice)
data(trial)
mice(trial, m = 2) %>%
complete() %>%
as_tibble() %>%
glm(death ~ marker grade, data = ., family = "binomial") %>%
tbl_regression(
tidy_fun = tidy_standardize,
exponentiate = TRUE, conf.int = TRUE, conf.level = 0.95
)
CodePudding user response:
Unfortunately, tidiers cannot be combined or stacked.
To get what you're after, I would recommend that you standardize the covariates in the modelling step, rather than waiting until the model has been estimated.
Example below!
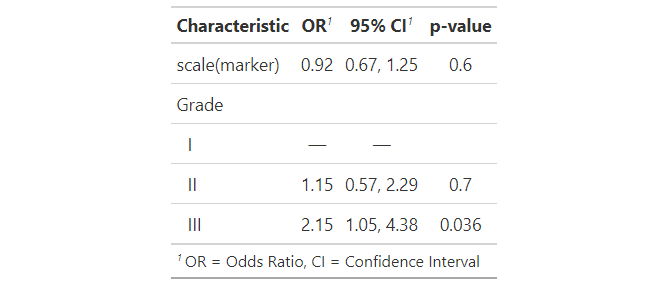
library(gtsummary)
packageVersion("gtsummary")
#> [1] '1.5.0'
tbl <-
suppressWarnings(mice::mice(trial, m = 2)) %>%
with(glm(death ~ scale(marker) grade, family = "binomial")) %>%
tbl_regression(exponentiate = TRUE)
#> pool_and_tidy_mice(): Tidying mice model with
#> `mice::pool(x) %>% mice::tidy(exponentiate = TRUE, conf.int = TRUE, conf.level = 0.95)`
 Created on 2021-12-10 by the reprex package (v2.0.1)
Created on 2021-12-10 by the reprex package (v2.0.1)
