Here is an example of my dataset:
df <- data.frame(
id = c(13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 62, 63, 64, 65, 13, 14, 15, 16, 17, 18,
19, 20, 21, 22, 23, 24, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 62, 63, 64, 65, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24,
29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 62, 63, 64, 65),
collection_point = c(rep(c("Baseline", "Immediate", "3M"), each=28)),
intervention = c(rep(c("B", "A", "C", "B", "C", "A", "A", "B", "A", "C", "B", "C",
"A", "A", "B", "A", "C", "B", "C", "A", "A"), each = 4)),
scale_A = c(6.5, 7.0, 6.25, 6.0, NA, 7.5, 7.5,
8.0, 7.5, 6.75, 7.5, 6.75, 6.75, 6.5,
5.75, 6.75, 7.75, 7.5, 7.75, 7.25, 7.75,
7.25, 7.25, 5.75, 6.75, NA, 6.75, 7.5,
6.75, 7.0, 6.5, 7.0, 7.5, 7.5, 7.5,
7.75, 7.25, 7.25, 7.25, 7.5, 6.5, 6.25,
6.25, 7.25, 7.5, 6.75, 7.25, 7.25, 7.5,
7.25, 7.5, 7.25, NA, 7.0, 7.5, 7.5,
6.75, 7.25, 6.5, 7.0, 7.5, 7.5, 7.5,
7.75, 7.5, 7.5, 7.5, 7.5, 6.5, 5.75,
6.25, 6.75, 7.5, 7.25, 7.25, 7.5, 7.75,
7.75, 7.75, 7.5, NA, NA, NA, NA))
where,
id = participant
collection_point = times data was collected from participant (repeated measure)
intervention = group each participant was randomized to (fixed effect)
scale_A = questionnaire score that each participant completed at each data collection point (outcome)
Participants were randomized to one of three interventions and completed the same scales (scales A-C) at three different time points to determine any improvements over time.
I have used the code
mixed.lmer.A1<-lmer(scale_A~intervention*collection_point (collection_point|id), control =
lmerControl(check.nobs.vs.nRE = "ignore"), data = df)
I can use plot_model(mixed.lmer.A1) and use the function terms to select only the interaction effects (ex: terms = c("interventionB:collection_point3M") to create a forest plot. However, I think it would look much neater to only have the interventions on the y axis and have multiple bands that represent each collection_point. Desired output like this:
Any idea how I can do this? Thanks!
CodePudding user response:
Here is one solution:
library(ggplot2)
library(lme4)
#> Loading required package: Matrix
library(sjPlot)
#> Install package "strengejacke" from GitHub (`devtools::install_github("strengejacke/strengejacke")`) to load all sj-packages at once!
df <- data.frame(
id = c(13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 62, 63, 64, 65, 13, 14, 15, 16, 17, 18,
19, 20, 21, 22, 23, 24, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 62, 63, 64, 65, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24,
29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 62, 63, 64, 65),
collection_point = c(rep(c("Baseline", "Immediate", "3M"), each=28)),
intervention = c(rep(c("B", "A", "C", "B", "C", "A", "A", "B", "A", "C", "B", "C",
"A", "A", "B", "A", "C", "B", "C", "A", "A"), each = 4)),
scale_A = c(6.5, 7.0, 6.25, 6.0, NA, 7.5, 7.5,
8.0, 7.5, 6.75, 7.5, 6.75, 6.75, 6.5,
5.75, 6.75, 7.75, 7.5, 7.75, 7.25, 7.75,
7.25, 7.25, 5.75, 6.75, NA, 6.75, 7.5,
6.75, 7.0, 6.5, 7.0, 7.5, 7.5, 7.5,
7.75, 7.25, 7.25, 7.25, 7.5, 6.5, 6.25,
6.25, 7.25, 7.5, 6.75, 7.25, 7.25, 7.5,
7.25, 7.5, 7.25, NA, 7.0, 7.5, 7.5,
6.75, 7.25, 6.5, 7.0, 7.5, 7.5, 7.5,
7.75, 7.5, 7.5, 7.5, 7.5, 6.5, 5.75,
6.25, 6.75, 7.5, 7.25, 7.25, 7.5, 7.75,
7.75, 7.75, 7.5, NA, NA, NA, NA))
mixed.lmer.A1 <- lmer(scale_A~intervention*collection_point (collection_point|id), control =
lmerControl(check.nobs.vs.nRE = "ignore"), data = df)
#> Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
#> unable to evaluate scaled gradient
#> Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
#> Model failed to converge: degenerate Hessian with 1 negative eigenvalues
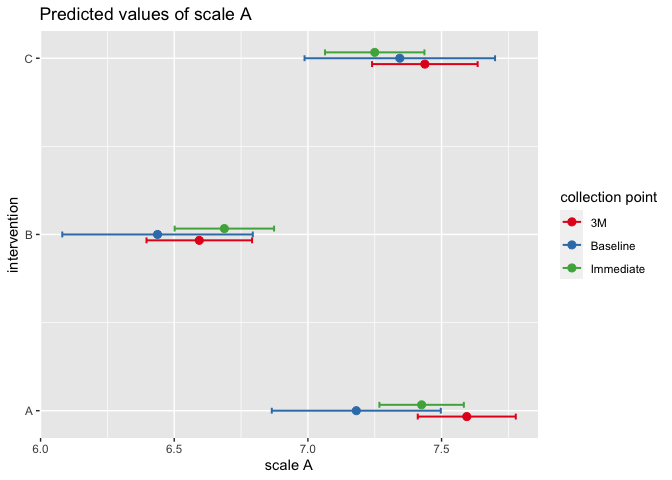
plot_model(mixed.lmer.A1, type = "int")
coord_flip()
Created on 2021-12-13 by the reprex package (v2.0.1)