I have imported data from excel (2 columns - Time and Voltage) into 2 NumPy arrays. The time column is imported as pandas timestamp data (<class 'pandas._libs.tslibs.timestamps.Timestamp'>) in the format:
yyyy-mm-dd HH:MM:SS.microseconds (Example below)
2021-12-29 18:39:29.845000
I am trying to display a plot with time on the x-axis and voltage on the y-axis. But when I plot it, the time is displayed as HH:MM:SS.
I want to display the time from 0 to 10 seconds since this is how long each experiment takes but there is no information on how to take an array with timestamp data and convert it to an array of decimal numbers going from 0.000 to 10.000 seconds, for example:
time = [0.000, 0.001, 0.002, ... , 9.998, 9.999, 10.000]
I have tried things like looping through the array and taking the difference from the first value but that takes 1hour since my data set is 500,000 points long. So even if that did work, I do not have the time to do it for all of the graphs I want to plot.
I have also tried getting the array with the time values and taking away the first value to get the time difference:
dateTimeDifference = time - time[0]
But i get this error:
TypeError: unsupported operand type(s) for -: 'numpy.ndarray' and 'Timestamp'
However, I have a feeling that even if i were to do this, i would get an array in the format of 'yyyy-mm-dd HH:MM:SS.microseconds' instead of what i want which is an array in the format 'SS.microseconds'.
Can you help?
CodePudding user response:
You can convert the pandas timestamps into time difference data:
import numpy as np
import pandas as pd
#create sample data
np.random.seed(123)
date_range = pd.date_range("2022-1-3 12:34:50", periods=50, freq="7 ms")
vals = np.random.randint(-10, 50, len(date_range))
df_exp = pd.DataFrame({"Exp_Time": date_range, "Exp_Values": vals})
#convert to seconds
#it is not necessary to store these values in an extra column
#you can also generate them on the fly in the plot
df_exp["Exp_MS"] = df_exp["Exp_Time"].diff().dt.microseconds.fillna(value=0).cumsum()/1000000
#plot
from matplotlib import pyplot as plt
plt.plot(df_exp["Exp_MS"], df_exp["Exp_Values"])
plt.xlabel("Time (in s)")
plt.show()
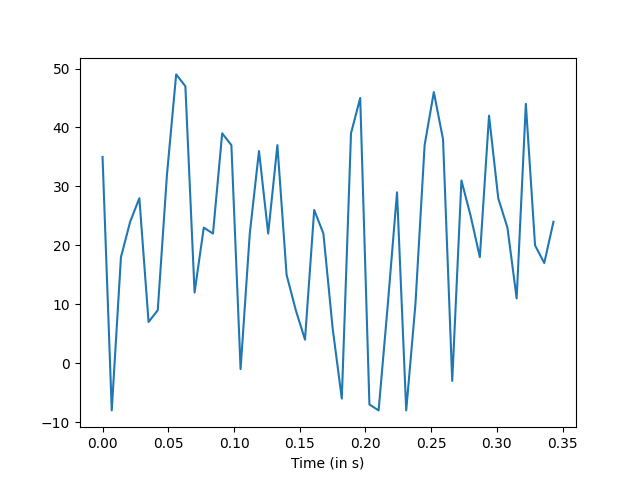
Sample output:
Some additional explanation - the conversion happens here:
df_exp["Exp_Time"].diff().dt.microseconds.fillna(value=0).cumsum()/1000000
This is just a series of chained operations:
- take the timestamp column
df_exp["Exp_Time"] - calculate the difference between values
.diff() - convert it to datetime microseconds
.dt.microseconds - set the first value to zero
.fillna(value=0) - create the cumulative sum of the time differences
.cumsum() - and finally transform the microsecond values into seconds
/1000000
Et voila, now you are ready to adapt this to your needs. As mentioned already, you could also simply plot the pandas series directly if you don't use these values again for something else:
plt.plot(df_exp["Exp_Time"].diff().dt.microseconds.fillna(value=0).cumsum()/1000000, df_exp["Exp_Values"])
CodePudding user response:
Try and time this code, where time is your time array. Assign it to new array and use that as your X-axis.
[i - time[0] for i in time]