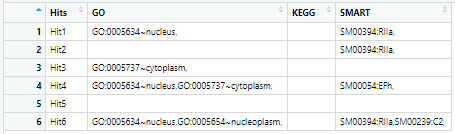
I have a dummy dataframe df which has dimensions 6 X 4.
df <- data.frame(
Hits = c("Hit1", "Hit2", "Hit3", "Hit4", "Hit5", "Hit6"),
GO = c("GO:0005634~nucleus,", "", "GO:0005737~cytoplasm,", "GO:0005634~nucleus,GO:0005737~cytoplasm,", "",
"GO:0005634~nucleus,GO:0005654~nucleoplasm,"),
KEGG = c("", "", "", "", "", ""),
SMART = c("SM00394:RIIa,", "SM00394:RIIa,", "", "SM00054:EFh,",
"", "SM00394:RIIa,SM00239:C2,"))
df looks like this
The elements in the columns consist of two parts:
- an
identifier(e.g.GO:0005634~,SM00394:etc.) - a
term(e.g.nucleus,EFhetc.)
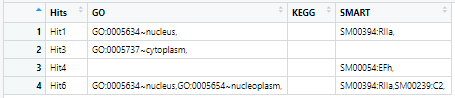
For each column I want to retain a row if it contains atleast one term which is not present in any row above it. e.g. in the column GO rows 1 and 3 contain unique terms, so these should be retained. Row 4 contains terms which are already present in rows 1 and 3, so it should be dropped. Row 6 has one term which is not present in any row above it, hence it should also be retained.
I have been able to come up with regular expressions to extract the terms from the columns GO and SMART
Regex for GO: (?<=~).*?(?=,(?:GO:\\d ~|$))
Regex for SMART: (?<=:).*?(?=,(?:\\w \\d :|$))
But I'm unable to figure out a way to integrate the regex and the conditions mentioned above into a solution. The output should look like this
Any suggestions on how to solve this?
CodePudding user response:
Here is a general approach that will handle GO, SMART, and potentially KEGG, though it is impossible to say without any information about KEGG.
The function f below takes as arguments
x, a character vectorsplit, the delimiter separating items in listssep, the delimiter separating identifiers and terms within items
and returns a logical vector indexing the elements of x with at least one non-duplicated term.
f <- function(x, split, sep) {
l1 <- strsplit(x, split)
tt <- sub(paste0("^[^", sep, "]*", sep), "", unlist(l1))
l2 <- relist(duplicated(tt), l1)
!vapply(l2, all, NA)
}
Applying f to GO and SMART:
nms <- c("GO", "SMART")
l <- Map(f, x = df[nms], split = ",", sep = c("~", ":"))
l
## $GO
## [1] TRUE FALSE TRUE FALSE FALSE TRUE
##
## $SMART
## [1] TRUE FALSE FALSE TRUE FALSE TRUE
Setting to "" elements of GO and SMART with zero non-duplicated terms, then filtering out empty rows, we obtain the desired result:
df2 <- df
df2[nms] <- Map(replace, df2[nms], lapply(l, `!`), "")
df2[Reduce(`|`, l), ]
## Hits GO KEGG SMART
## 1 Hit1 GO:0005634~nucleus, SM00394:RIIa,
## 3 Hit3 GO:0005737~cytoplasm,
## 4 Hit4 SM00054:EFh,
## 6 Hit6 GO:0005634~nucleus,GO:0005654~nucleoplasm, SM00394:RIIa,SM00239:C2,
CodePudding user response:
The following algorithm is applied to each term (GO, SMART, KEGG):
- extract the identifier term list as comma-separated. See
stringr::str_splitetc. - extract the term as regex
- cumulate all the terms along the dataframe as they appear
- extract the difference between each row and the row immediately preceding
- replace the string with
""if no new term is introduced - filter rows where not all the terms are
""
library(dplyr)
library(stringr)
library(purrr)
termred <- function(terms, rx) {
terms |>
stringr::str_split(",") |>
purrr::map(stringr::str_trim) |>
purrr::map(~{.x[.x != ""]}) |>
purrr::map(~stringr::str_extract(.x, rx)) |>
purrr::accumulate(union) %>%
{mapply(setdiff, ., lag(., 1), SIMPLIFY = TRUE)} %>%
{ifelse(sapply(., length) > 0, terms, "")}
}
df |>
transform(GO = termred(GO, "~.*$")) |>
transform(SMART = termred(SMART, ":.*$")) |>
filter(GO != "" | SMART != ""| KEGG != "")
##> Hits GO KEGG SMART
##>1 Hit1 GO:0005634~nucleus, SM00394:RIIa,
##>2 Hit3 GO:0005737~cytoplasm,
##>3 Hit4 SM00054:EFh,
##>4 Hit6 GO:0005634~nucleus,GO:0005654~nucleoplasm, SM00394:RIIa,SM00239:C2,