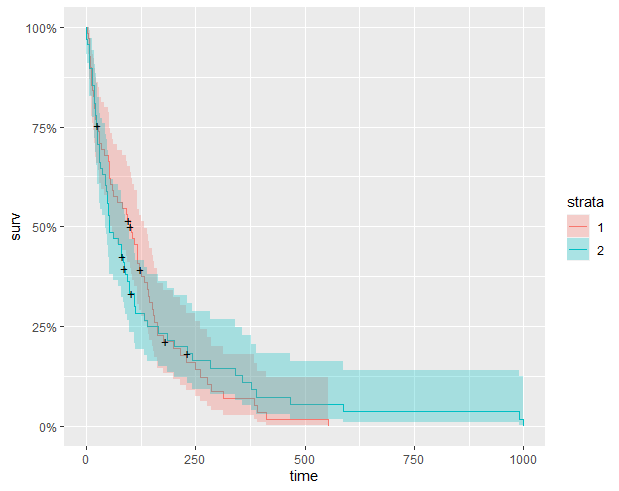
I am generating kaplan-meier plots within R and I was wondering if there were a way to create an interactive kaplan-meier plot using the plotly package? Ideally, I would like to be able to trace the survival "line" using my mouse cursor with the plotly tooltip box telling me the group, the survival rate, and the time as well.
Here is an mock example of survival analysis that I would like to recreate within plotly:-
library(survival)
library(ranger)
library(ggplot2)
library(dplyr)
library(ggfortify)
library(plotly)
#------------
data(veteran)
head(veteran)
km_trt_fit <- survfit(Surv(time, status) ~ trt, data=veteran)
#using autoplot to create graph
autoplot(km_trt_fit)
But when I try to use plotly, I get this error:-
ggplotly(km_trt_fit)
#Error in UseMethod("ggplotly", p) :
#no applicable method for 'ggplotly' applied to an object of class "survfit"
What would be the additional steps I need to take to get the km_trt_fit variable to be amenable to the ggplotly function? And would I be able to see within the tooltip the features which I have outlined above? Many thanks :)
CodePudding user response:
One option is to extract the ggplot figure from a ggsurvplot object.
library(survminer)
p <- ggsurvplot(km_trt_fit)
ggplotly(p[[1]])
To obtain table data on events at different time points, you can work with p[[3]].