I'm working with the cake dataset and constructed the following model
mod <- lmer(angle ~ recipe*temp (1|replicate), data=cake)
newdat <- expand.grid(recipe=unique(cake$recipe),
temp=c(min(cake$temp),
max(cake$temp)))
Then I wanted to plot the regression lines for each replicate separately for each recipe (A, B and C)
ggplot(cake, aes(x=temp, y=angle, color=recipe))
geom_line(aes(y=predict(mod), group=replicate, size="Replicate"))
geom_line(data=newdat, aes(y=predict(mod, re.form=~0, newdata=newdat), size="Population"))
scale_size_manual(name="Predictions", values=c("Replicate"=0.5, "Population"=1))
I have two questions. First, it seems that Population lines B and C are not parallel, even though I haven't defined a random slope model.
Second, I cannot make the code above to plot the regression lines for each replicate split by recipe.
CodePudding user response:
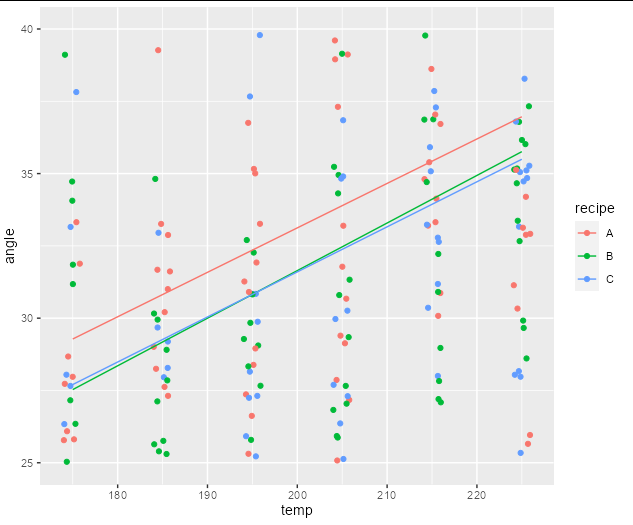
To answer your first question, your lines are not parallel because you have an interaction term between recipe and temp in your formula. You therefore have different slopes for the fixed effects (i.e. the change in angle per unit change in temp is dependent on recipe). We can see this even if we plot a simple lm:
library(ggplot2)
mod <- lm(angle ~ recipe * temp, data = cake)
newdat <- expand.grid(recipe=unique(cake$recipe),
temp=c(min(cake$temp),
max(cake$temp)))
newdat$angle <- predict(mod, newdata = newdat)
ggplot(cake, aes(temp, angle, color = recipe))
geom_point(position = position_jitter(width = 1))
geom_line(data = newdat)
scale_y_continuous(limits = c(25, 40))
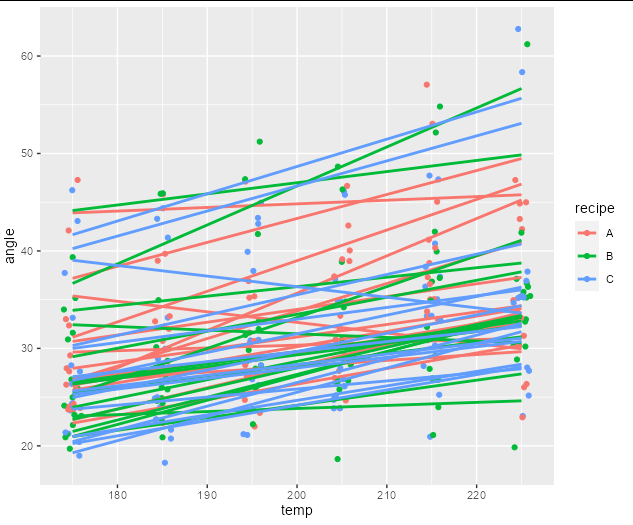
To answer the second question, you need to group by the interaction of replicate and recipe to get correctly colored lines for each of the replicates. You also need to have individual lm generated lines for them. The easiest way to do this is via geom_smooth with method = lm:
ggplot(cake, aes(temp, angle, color = recipe))
geom_point(position = position_jitter(width = 1))
geom_smooth(aes(group = interaction(replicate, recipe)), se = FALSE, method = lm)