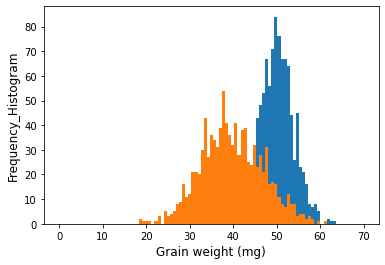
I have a question about overlapping graphs inside one panel in Python.
I generated numbers of two groups
import numpy as np
cv1 = np.random.normal(50, 4, 1000)
cv2 = np.random.normal(40, 7, 1000)
import matplotlib.pyplot as plt
plt.hist(cv1, numpy.linspace(0, 70, 100))
plt.hist(cv2, numpy.linspace(0, 70, 100))
plt.xlabel("Grain weight (mg)", size=12)
plt.ylabel("Frequency_Histogram", size=12)
plt.show()
It works perfectly. However, if the data is the format of data frame, how can I make the same graph?
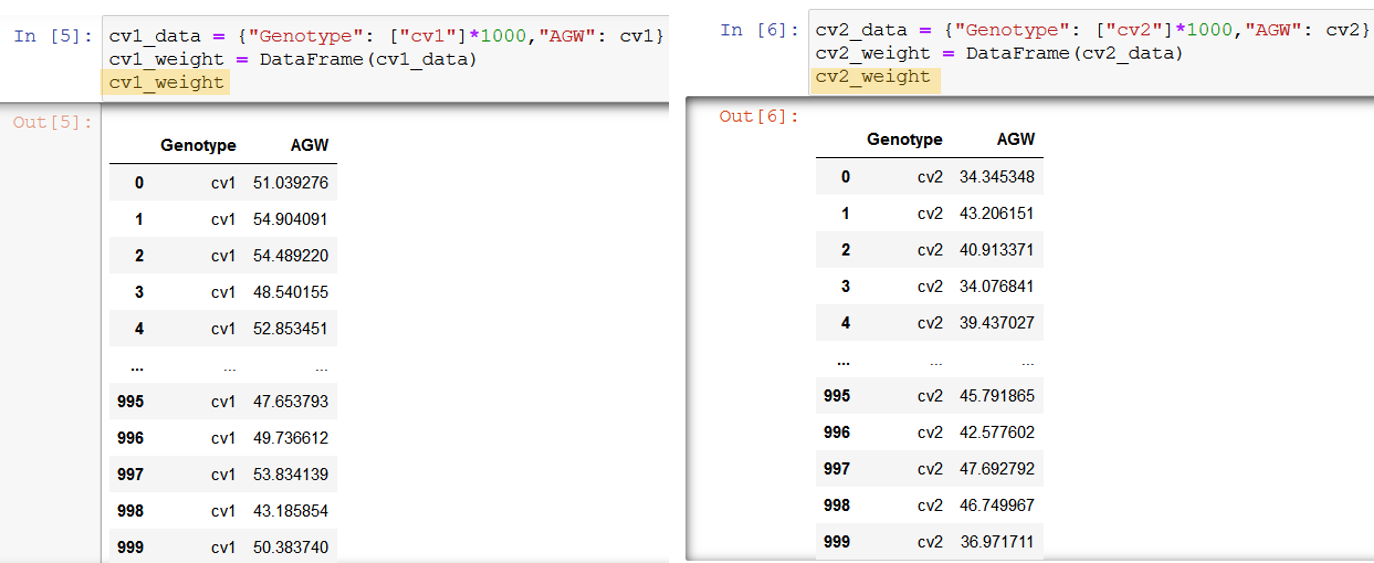
For example, I simulated that I have two data set for different genotypes, cv1, cv2. In each data, grain weight for 1,000 grains was measured.
cv1_data = {"Genotype": ["cv1"]*1000,"AGW": cv1}
cv1_weight = DataFrame(cv1_data)
cv2_data = {"Genotype": ["cv2"]*1000,"AGW": cv2}
cv2_weight = DataFrame(cv2_data)
Then, I'd like to make the same graph above. So I tried below codes
plt.hist(cv1,y="AGW", numpy.linspace(0, 70, 100))
plt.hist(cv2,y="AGW", numpy.linspace(0, 70, 100))
plt.xlabel("Grain weight (mg)", size=12)
plt.ylabel("Frequency_Histogram", size=12)
plt.show()
but it does not work. Could you let me know how to make the same overlapping graph in case of data frame?
Always, many thanks!!!
CodePudding user response:
You can do that using Seaborn library in python
import numpy as np
import pandas as pd
import seaborn as sns
cv1 = np.random.normal(50, 4, 1000)
cv2 = np.random.normal(40, 7, 1000)
cv1_data = {"Genotype": ["cv1"]*1000,"AGW": cv1}
cv1_weight = pd.DataFrame(cv1_data)
cv2_data = {"Genotype": ["cv2"]*1000,"AGW": cv2}
cv2_weight = pd.DataFrame(cv2_data)
df = pd.concat([cv1_weight,cv2_weight],axis = 1)
df.columns = ['Genotype', 'cv1_AGW', 'Genotype', 'cv2_AGW']
sns.histplot(data = df[['cv1_AGW','cv2_AGW']])