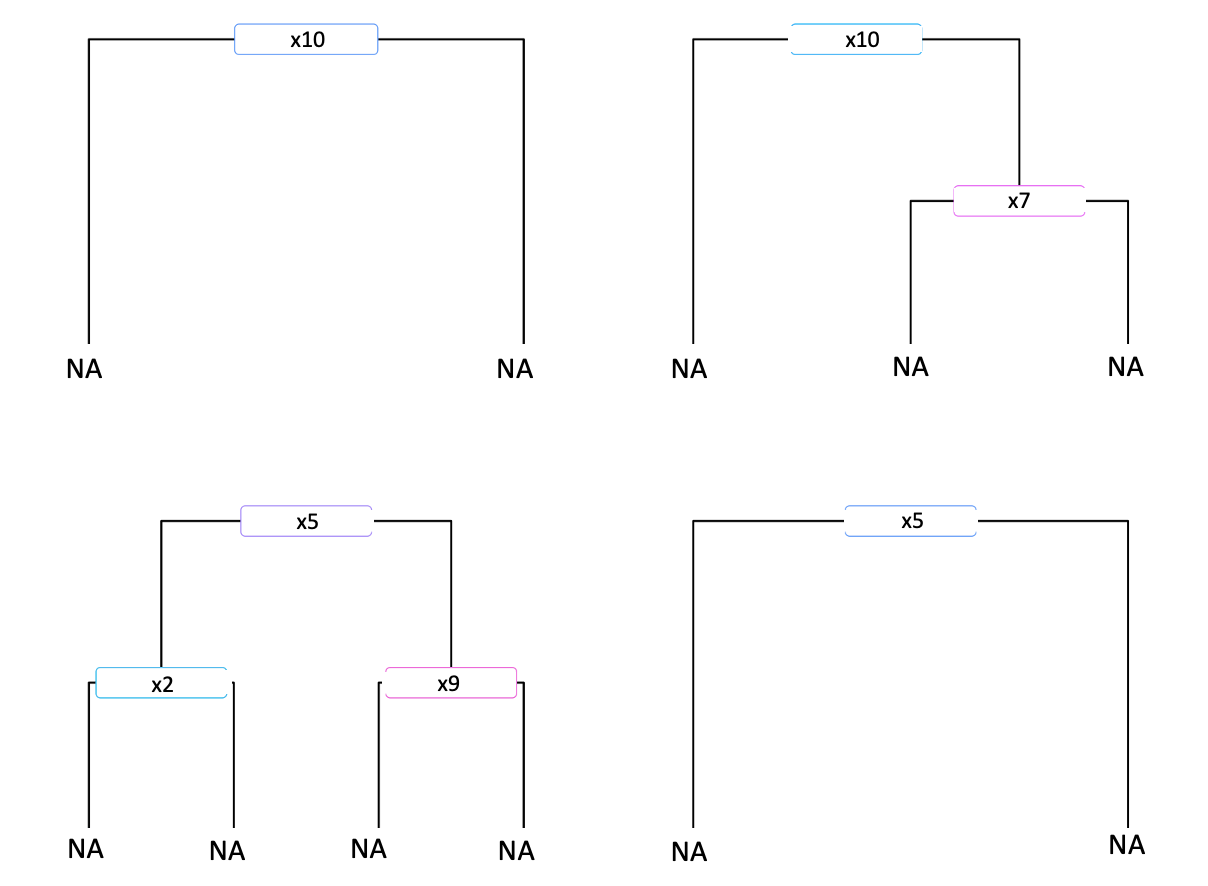
I'm trying to count each unique split rule from a data frame of decision trees in R. For example, if I have a data frame containing 4 trees like the one shown below:
df <- data.frame(
var = c('x10', NA, NA,
'x10', NA, 'x7', NA, NA,
'x5', 'x2', NA, NA, 'x9', NA, NA,
'x5', NA, NA),
num = c(1,1,1,
2,2,2,2,2,
1,1,1,1,1,1,1,
2,2,2),
iter = c(rep(1, 8), rep(2, 10))
)
> df
var num iter
1 x10 1 1
2 <NA> 1 1
3 <NA> 1 1
4 x10 2 1
5 <NA> 2 1
6 x7 2 1
7 <NA> 2 1
8 <NA> 2 1
9 x5 1 2
10 x2 1 2
11 <NA> 1 2
12 <NA> 1 2
13 x9 1 2
14 <NA> 1 2
15 <NA> 1 2
16 x5 2 2
17 <NA> 2 2
18 <NA> 2 2
The var column contains the variable name used in the splitting rule and is ordered by depth first. So, for example, the 4 trees created from that data would look like this:
I'm trying to find a way to return the count of each pair of variables used in a split rule, but grouped by iter. For example, if we look at the 2nd tree (i.e.,num == 2, iter == 1) we can see that x7 splits on x10. so, the pair x10:x7 appears 1 time when iter == 1.
My desired output would look something like this:
allSplits count iter
1 x10:x7 1 1
2 x5:x2 1 2
3 x5:x9 1 2
Any suggestions as to how I could do this?
CodePudding user response:
There is probably a package that knows how to operate on this kind of data frame, but maybe these two hand-crafted recursive functions can get you started.
mkTree <- function(x, pos = 1L) {
var <- x[pos]
if (is.na(var)) {
list(NA_character_, NULL, NULL, 1L)
} else {
node <- vector("list", 4L)
node[[1L]] <- var
node[[2L]] <- l <- Recall(x, pos 1L)
node[[3L]] <- r <- Recall(x, pos 1L l[[4L]])
node[[4L]] <- 1L l[[4L]] r[[4L]]
node
}
}
tabTree <- function(tree, sep = ":") {
x <- rep.int(NA_character_, tree[[4L]])
pos <- 1L
recurse <- function(subtree) {
var1 <- subtree[[1L]]
if (!is.na(var1)) {
for (i in 2:3) {
var2 <- subtree[[c(i, 1L)]]
if (!is.na(var2)) {
x[pos] <<- paste0(var1, sep, var2)
pos <<- pos 1L
Recall(subtree[[i]])
}
}
}
}
recurse(tree)
x <- x[!is.na(x)]
if (length(x)) {
x <- factor(x)
setNames(tabulate(x), levels(x))
} else {
integer(0L)
}
}
mkTree transforms into recursive lists the segments of var in your data frame that specify a tree. Nodes in these recursive structures have the form:
list(variable_name, left_node, right_node, subtree_size)
tabTree takes the mkTree result and returns a named integer vector tabulating the splits. So you could do
f <- function(x) tabTree(mkTree(x))
L <- tapply(df[["var"]], df[c("num", "iter")], f, simplify = FALSE)
to get a list matrix storing the tabulated splits for each [num, iter] pair (i.e., for each tree).
L
## iter
## num 1 2
## 1 integer,0 integer,2
## 2 1 integer,0
L[2L, 1L]
## [[1]]
## x10:x7
## 1
L[1L, 2L]
## [[1]]
## x5:x2 x5:x9
## 1 1
And you could sum over num to get tabulated splits for each level of iter.
g <- function(l) {
x <- unlist(unname(l))
tapply(x, names(x), sum)
}
apply(L, 2L, g)
## $`1`
## x10:x7
## 1
## $`2`
## x5:x2 x5:x9
## 1 1