I'm trying to plot the coefficients of three zero-inflated negative binomial models.
My data looks like this:
# A tibble: 6 x 8
Park Week Coy_Season Reports_per_week Number_AC Reports_month_pri~ Number_4w_AC Year_numeric
<chr> <date> <chr> <dbl> <int> <dbl> <int> <dbl>
1 14st NE - Covent~ 2018-04-29 1 0 0 0 0 1
2 14st NE - Covent~ 2018-05-06 2 0 0 0 0 1
3 Airways Park 2021-07-18 2 3 0 0 0 4
4 Aspen Heights 2021-03-28 1 2 12 0 0 4
5 Aspen Heights 2021-05-09 2 0 13 0 0 4
Where Coy_Season vary between 1-3 and Year_numeric varies between 1-4.
zero.infl.neg.bin.mod.1 <- glmmTMB(Reports_per_week ~ (1|Park) Reports_month_prior,
ziformula = ~ Reports_month_prior Number_4w_AC (1|Park)
Year_numeric factor(Coy_Season),
data = Reports_per_park_per_week_3,
family = nbinom2, na.action = "na.fail")
zero.infl.neg.bin.mod.2 <- glmmTMB(Reports_per_week ~ (1|Park) Reports_month_prior Year_numeric,
ziformula = ~ Reports_month_prior Number_4w_AC (1|Park)
Year_numeric factor(Coy_Season),
data = Reports_per_park_per_week_3,
family = nbinom2, na.action = "na.fail")
zero.infl.neg.bin.mod.3 <- glmmTMB(Reports_per_week ~ (1|Park) Reports_month_prior Number_4w_AC,
ziformula = ~ Reports_month_prior Number_4w_AC (1|Park)
Year_numeric factor(Coy_Season),
data = Reports_per_park_per_week_3,
family = nbinom2, na.action = "na.fail")
library(broom)
library(broom.mixed)
library(dotwhisker)
dwplot(list(zero.infl.neg.bin.mod.1, zero.infl.neg.bin.mod.2, zero.infl.neg.bin.mod.3),
effects = "fixed")
The resulting plot merges my coefficients from my conditional model and my zero-inflated model when a variable is present in both models, which is incorrect. How can I make my dwplot model the variables in my conditional models independently from those in my zero-inflated models (one coefficient for each conditional model, and one coefficient for each ZI model)?
CodePudding user response:
Load packages:
library(glmmTMB)
library(broom.mixed)
library(tidyverse)
library(dotwhisker)
theme_set(theme_bw())
Fit models (reproducible example):
m1 <- glmmTMB(count ~ spp mined (1|site),
zi=~spp mined,
family=nbinom2, data=Salamanders)
m2 <- update(m1, . ~ . - (1|site))
Put together combined, tidied results:
tt <- (list(with_RE = m1, no_RE = m2)
%>% purrr::map_dfr(tidy, effects = "fixed", conf.int = TRUE,
.id = "model")
%>% select(model, component, term, estimate, conf.low, conf.high)
## drop conditional intercept term (not interesting)
%>% filter(!(term == "(Intercept)" & component == "cond"))
## create new 'term' that combines component and term
%>% mutate(term_orig = term,
term = forcats::fct_inorder(paste(term, component, sep = "_")))
)
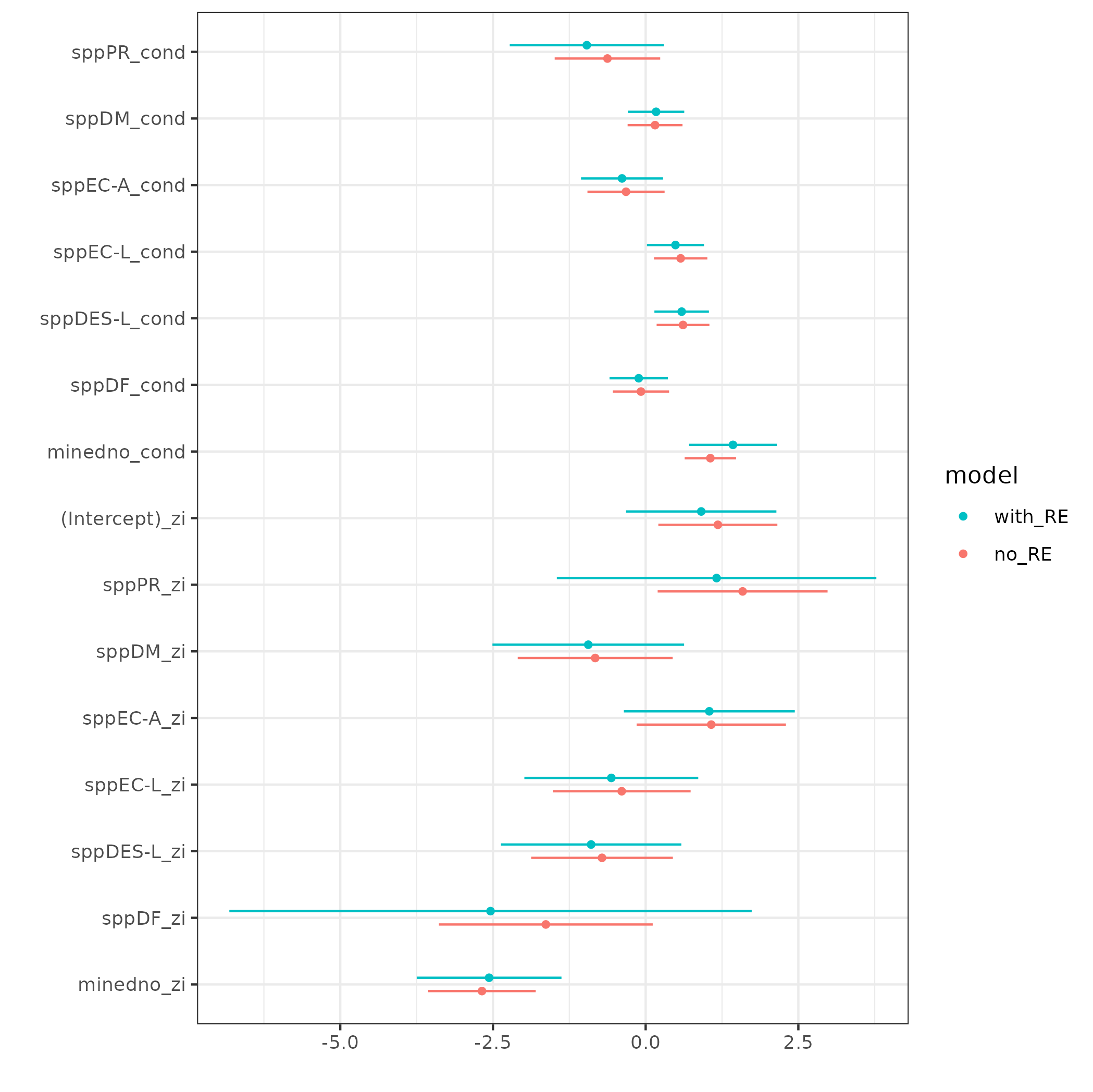
Now that the term values are unique, this can be plugged into dwplot:
dwplot(tt)
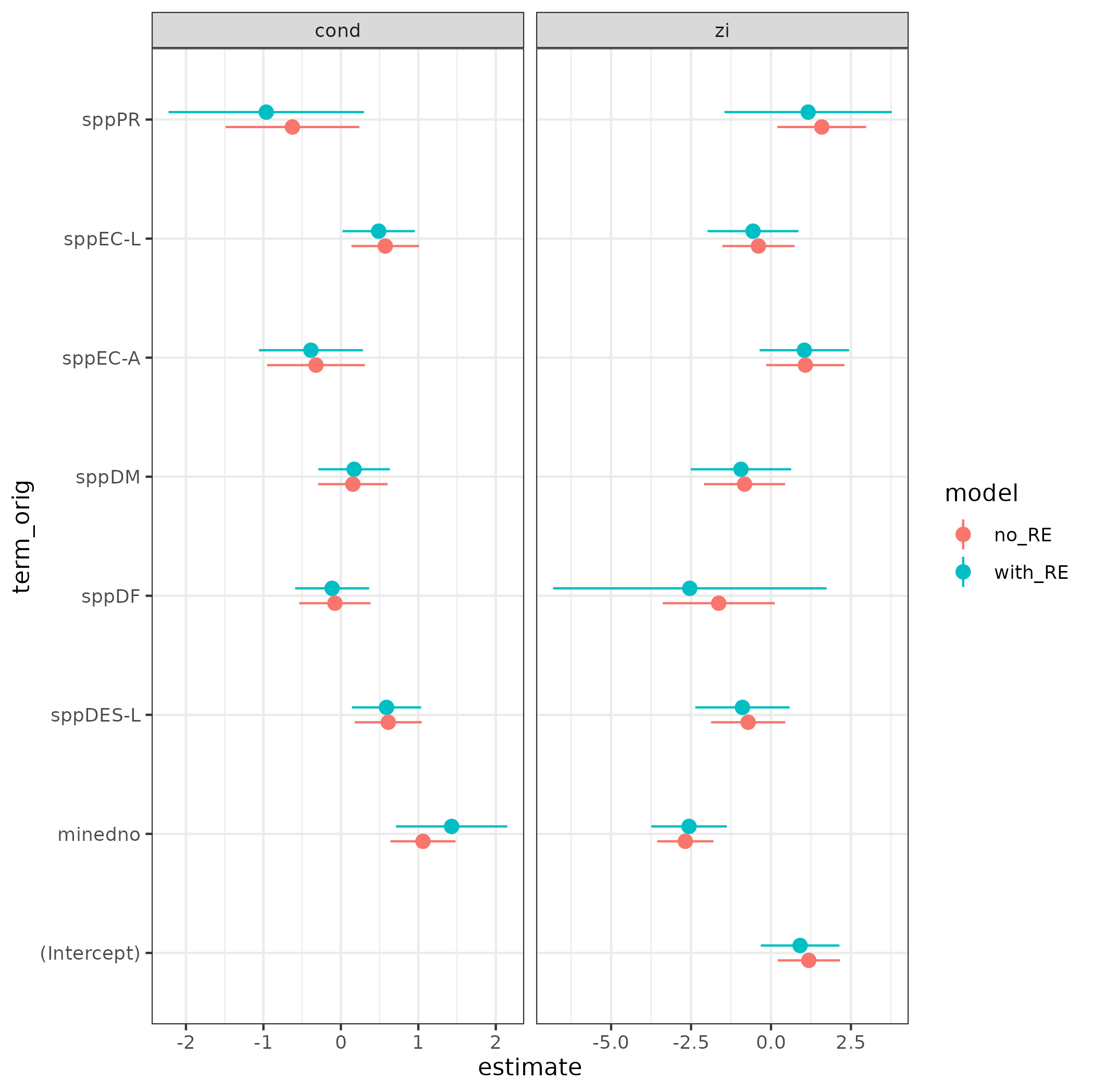
Or use ggplot geom_pointrange for more flexibility:
(ggplot(tt)
aes(x = estimate, xmin = conf.low, xmax = conf.high, y = term_orig)
geom_pointrange(aes(colour = model),
position = position_dodge(width = 0.25))
facet_wrap(~component, scale = "free_x")
)