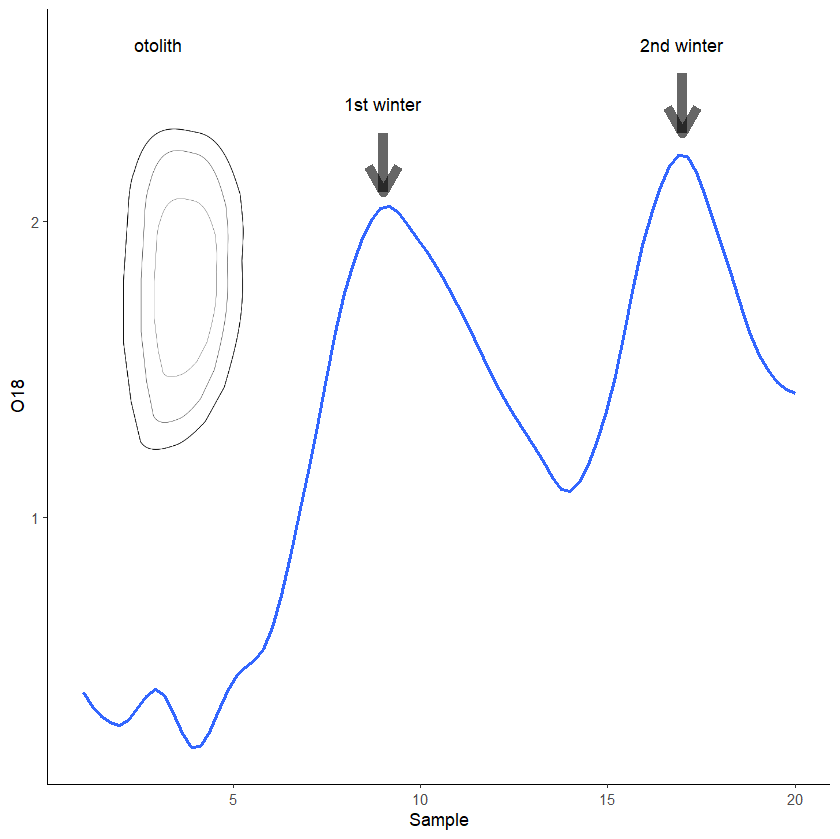
I am working on a plot that will have the data curve, a png image inset, as well as annotations. However, every time I add my inset image all of my annotations disappear and I'm not sure what I am doing wrong. Help would be greatly appreciated! I still need to work on my axis labels and such but am stuck here! Additionally, if anyone has any style tips or recommendations I am all ears!
o18 <- data.frame(Sample=1:20, O18 = c(.41,.3, .42,.22, .45,.61, 1.15, 1.77,2.05,
1.93,1.72,1.46,1.25, 1.09,1.38,1.96, 2.23,
1.93, 1.56,1.42))
library(png)
oto_ill <- readPNG('otolith_illustration.png', native = TRUE)
library(patchwork)
o18.plot2 <- ggplot(data=o18, aes(x=Sample, y = O18))
geom_line(colour='NA')
theme_classic()
geom_smooth(method="loess", span=0.2)
inset_element(oto_ill, left =0.01, right =0.3, bottom = 0.4, top =1)
annotate(geom="text", x=3, y=2.6, label="otolith", size=4, color='black',
parse=TRUE)
annotate(geom="text", x=9, y=2.4, label="1st winter" , size=4, color='black',
parse=FALSE)
annotate(geom="text", x=17, y=2.6, label="2nd winter", size=4, color='black',
parse=FALSE)
annotate("segment", x = 9, xend = 9, y = 2.3, yend = 2.1, colour = "black", size=3, alpha=0.6, arrow=arrow())
annotate("segment", x = 17, xend = 17, y = 2.5, yend = 2.3, colour = "black", size=3, alpha=0.6, arrow=arrow())
o18.plot2
CodePudding user response:
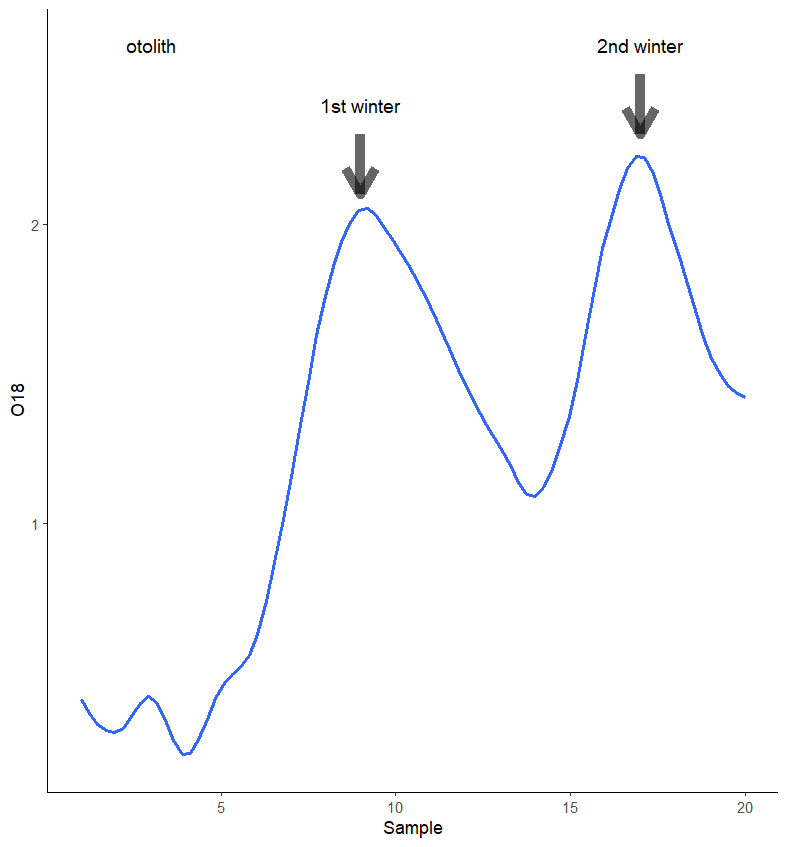
You can use annotate_custom instead of inset_element.
library(png)
library(ggplot2)
o18 <- data.frame(Sample=1:20,
O18 = c(.41,.3, .42,.22, .45,.61, 1.15, 1.77,2.05,
1.93,1.72,1.46,1.25,1.09,1.38,1.96, 2.23,
1.93, 1.56,1.42))
oto_ill <- readPNG('otolith_illustration.png', native = TRUE)
oto_grob <- grid::rasterGrob(oto_ill, interpolate=TRUE)
o18.plot2 <- ggplot(data=o18, aes(x=Sample, y = O18))
geom_line(colour='NA')
geom_smooth(method="loess", span=0.2)
annotate(geom="text", x=3, y=2.6, label="otolith", size=4, color='black',
parse=TRUE)
annotate(geom="text", x=9, y=2.4, label="1st winter" , size=4, color='black',
parse=FALSE)
annotate(geom="text", x=17, y=2.6, label="2nd winter", size=4, color='black',
parse=FALSE)
annotate("segment", x = 9, xend = 9, y = 2.3, yend = 2.1, colour = "black", size=3, alpha=0.6, arrow=arrow())
annotate("segment", x = 17, xend = 17, y = 2.5, yend = 2.3, colour = "black", size=3, alpha=0.6, arrow=arrow())
annotation_custom(oto_grob, xmin=1, xmax=6, ymin=0, ymax=3.5)
theme_classic()
o18.plot2
CodePudding user response:
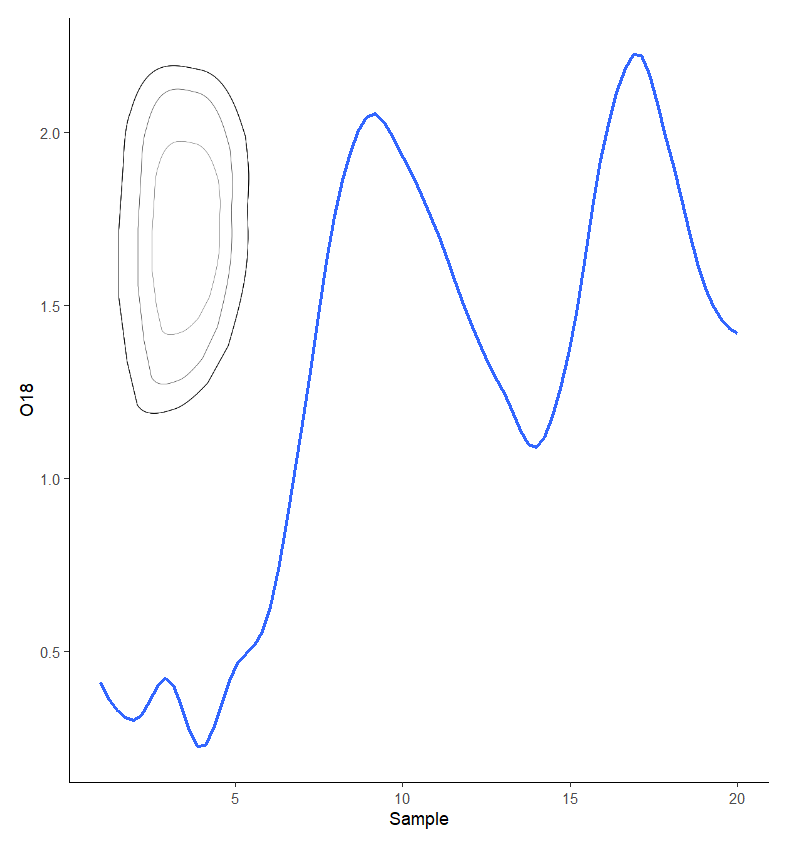
The following code worked, using patchwork and moving the oto_grob to the final line of code for the plot.
library(png)
oto_ill <- readPNG('otolith_illustration.png', native = TRUE)
oto_grob <- grid::rasterGrob(oto_ill, interpolate=TRUE)
#make the plot with annotations
library(patchwork)
o18
o18.plot2 <- ggplot(data=o18, aes(x=Distance, y = O18))
geom_line(colour='NA')
labs(y=expression(delta^18*O), x = "Distance from Core (mm)")
theme_classic()
geom_smooth(method="loess", span=0.2)
annotate(geom="text", x=.19, y=2.49, label="otolith", size=4,
color='black',parse=TRUE)
annotate(geom="text", x=.35, y=2.4, label="1st winter" , size=4,
color='black',parse=FALSE)
annotate(geom="text", x=.57, y=2.6, label="2nd winter", size=4,
color='black',parse=FALSE)
annotate("segment", x = .35, xend = .35, y = 2.3, yend = 2.1,
colour = "black", size=3, alpha=0.8, arrow=arrow())
annotate("segment", x = .57, xend = .57, y = 2.5, yend = 2.3,
colour = "black", size=3, alpha=0.8, arrow=arrow())
annotate(geom="text", x=.15, y=.05, label="larval stage", size=4,
color='black',parse=FALSE)
annotate(geom="text", x=.663, y=.05, label="catch", size=4,
color='black',parse=FALSE)
annotate("segment", x = .2, xend = .63, y = .05, yend = .05,
colour = "black", size=3, alpha=0.8, arrow=arrow())
inset_element(oto_grob, left =0.068, right =0.35, bottom = 0.4, top =.9,
align_to = 'full')
o18.plot2