I have a dataframe on R similar to this one, only it is 2000 rows long. Throughout the dataframe I have this alternation of SEQ1 and SEQ2 within a single read called "id read". These sequences alternate, and SEQ1 is always 1 nucleotide away from SEQ1, while SEQ2 from SEQ1 about 335 nucleotides, sometimes jumps and goes to 670. The sequences are both in forward and in revers, as can be seen from the value of the end coordinate which is sometimes less than the start coordinate.
| sequence | id read | start | end | sequencedistance | sequencelength |
|---|---|---|---|---|---|
| SEQ1 | id read | 90 | 105 | 1 | 15 |
| SEQ2 | id read | 440 | 458 | 335 | 18 |
| SEQ1 | id read | 459 | 474 | 1 | 15 |
| SEQ2 | id read | 808 | 826 | 334 | 18 |
| SEQ1 | id read | 827 | 812 | 1 | 15 |
| SEQ2 | id read | 1148 | 1156 | 336 | 18 |
| SEQ1 | id read | 1157 | 1172 | 1 | 15 |
| SEQ2 | id read | 1850 | 1868 | 678 | 18 |
| SEQ1 | id read | 1869 | 1854 | 1 | 15 |
| SEQ2 | id read | 2187 | 2205 | 333 | 18 |
| SEQ1 | id read | 2206 | 2221 | 1 | 15 |
| SEQ2 | id read | 2887 | 2905 | 666 | 18 |
Would anyone have any ideas on how to plot this data and visually show the pattern that these sequences have within a read? I have tried plotting with horizontal lines, lollipop, point, but none of these methods are effective in representing the amount of data I have and to visually understand the behavior of these sequences. Would anyone have an idea of how to plot the pattern? If I wanted, I could also plot only a part of the large dataframe I have, but at least I would like to understand the particularity of these sequences in the ultra-long read taken into consideration.
CodePudding user response:
I'm still not exactly sure what you are looking for, but if every row i where sequence == "SEQ" has a paired row i 1 where sequence == "SEQ2", you can calculate the relative start and ends sites and then try to visualise it.
Assuming your data is in a variable called df, you can calculate these as follows.
df <- transform(
df,
rel_start = ifelse(
as.character(sequence) == "SEQ1",
start - start,
start - c(0, head(start, -1))
),
rel_end = ifelse(
as.character(sequence) == "SEQ1",
end - start,
end - c(0, head(start, -1))
)
)
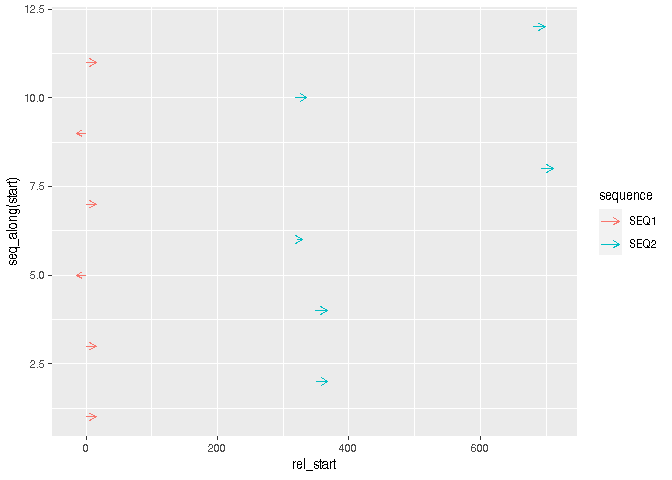
Then for visualisation, you can just use geom_segment(). You could use arrows to indicate the direction of the reads.
library(ggplot2)
ggplot(df, aes(rel_start, y = seq_along(start), colour = sequence))
geom_segment(aes(xend = rel_end, yend = seq_along(start)),
arrow = arrow(length = unit(2, "mm")))
Data loading:
txt <- "sequence id read start end sequencedistance sequencelength
SEQ1 id read 90 105 1 15
SEQ2 id read 440 458 335 18
SEQ1 id read 459 474 1 15
SEQ2 id read 808 826 334 18
SEQ1 id read 827 812 1 15
SEQ2 id read 1148 1156 336 18
SEQ1 id read 1157 1172 1 15
SEQ2 id read 1850 1868 678 18
SEQ1 id read 1869 1854 1 15
SEQ2 id read 2187 2205 333 18
SEQ1 id read 2206 2221 1 15
SEQ2 id read 2887 2905 666 18"
df <- read.table(text = txt, header = TRUE)
