We are studying different types of clusterization in R, and for the final task we need to show different graphs together. The ones I chose seem to be of different format, and I tried to look around for possible solutions to show them together, but came up short, probably due to my lack of experience (tried a bunch of options I found in similar questions, but nothing quite worked for me, or maybe I did something wrong).
I understand that there are roundabout ways to do this (which, according to some people, are way less troublesome too), and the prof is actually fine with me choosing only the graphs that are easily joined together, but at this point I want to satisfy my idle curiosity and ask whether there's a way to combine my graphs in R. I apologize if this was answered before, but I would be very grateful for assistance.
The graphs I randomly chose are:
clust1 = autoplot(fanny(xo, 5))
clust2 = autoplot(x_pca, data = x, colour = "region", loadings = TRUE, loadings.label = TRUE, frame = TRUE, frame.type = "norm")
clust3 = rpart.plot(x_tree)
clust4 = plot(as.phylo(x_clust), type = "fan", tip.color = colors(ct))
The first two I easily combined with grid.arrange, but both trees are giving me trouble. Thank you in advance for any help!
CodePudding user response:
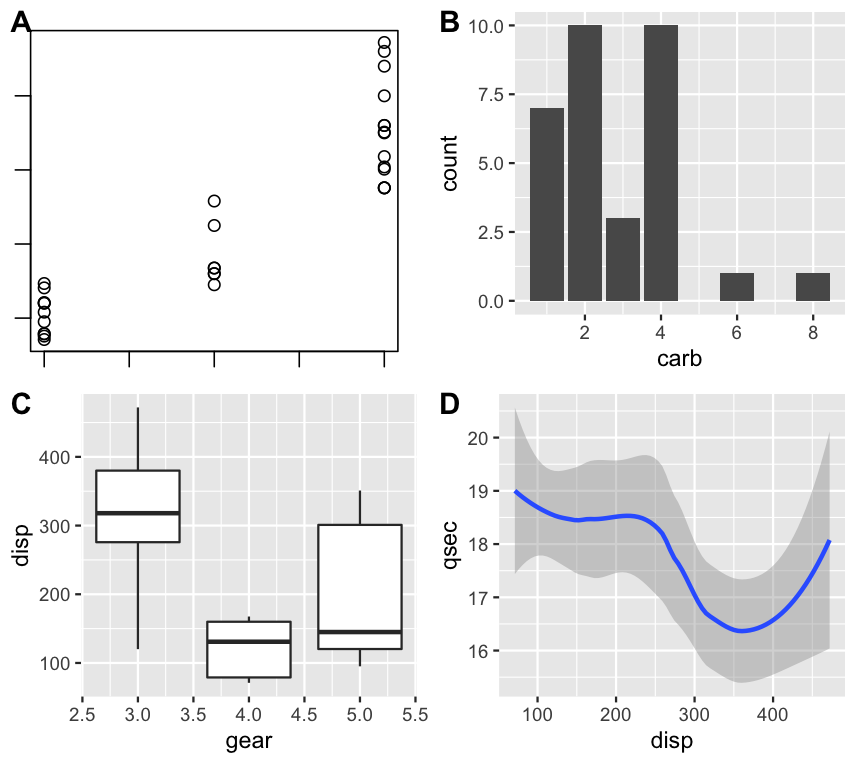
You could use the package cowplot which is very handy to combine multiple plots from base R and ggplot. Use the recordPlot function after your r base plot and use the plot_grid function to combine all the plots. Here is an example using the mtcars dataset:
library(ggplot2)
library(cowplot)
library(gridGraphics)
plot(mtcars$cyl, mtcars$disp)
p1 <- recordPlot()
p2 <- ggplot(mtcars) geom_boxplot(aes(gear, disp, group = gear))
p3 <- ggplot(mtcars) geom_smooth(aes(disp, qsec))
p4 <- ggplot(mtcars) geom_bar(aes(carb))
plot_grid(p1, p4, p2, p3, labels = 'AUTO')
Output:
CodePudding user response:
Here is a way to combine base R graphics with ggplot2 graphics. It is directly inspired in this answer by user Ricardo Saporta.
It uses grid graphics to create 2 view ports and prints the ggplot2 plots in those view ports.
library(ggplot2)
library(ggfortify)
library(rpart.plot)
#> Loading required package: rpart
library(cluster)
library(ape)
library(grid)
data(ruspini, package = "cluster")
data(ptitanic, package = "rpart.plot")
data(bird.orders, package = "ape")
old_par <- par(mfrow = c(2, 2))
vp.BottomLeft <- viewport(height=unit(.5, "npc"), width=unit(0.5, "npc"),
just=c("right","top"),
y=0.5, x=0.5)
vp.BottomRight <- viewport(height=unit(.5, "npc"), width=unit(0.5, "npc"),
just=c("left","top"),
y=0.5, x=0.5)
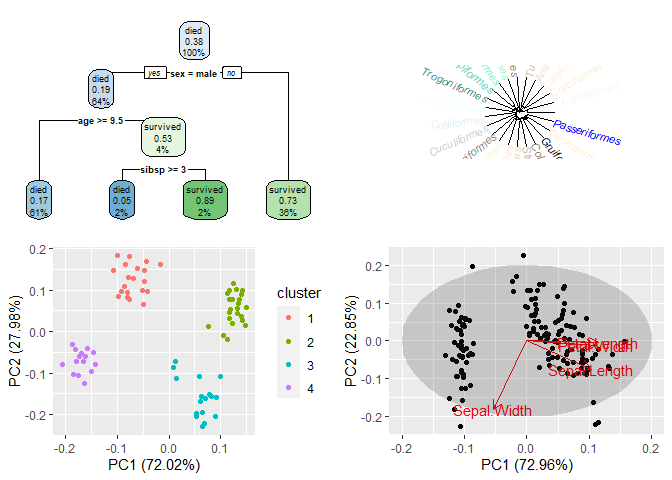
clust1 <- autoplot(fanny(ruspini, 4))
x_pca <- prcomp(iris[1:4], scale. = TRUE)
clust2 <- autoplot(x_pca, data = iris, loadings = TRUE, loadings.label = TRUE, frame = TRUE, frame.type = "norm")
binary.model <- rpart(survived ~ ., data = ptitanic, cp = .02)
clust3 <- rpart.plot(binary.model)
hc <- as.hclust(bird.orders)
ct <- length(hc$labels)
clust4 <- plot(as.phylo(hc), type = "fan", tip.color = colors(ct))
print(clust1, vp = vp.BottomLeft)
print(clust2, vp = vp.BottomRight)
par(old_par)
Created on 2022-03-19 by the reprex package (v2.0.1)