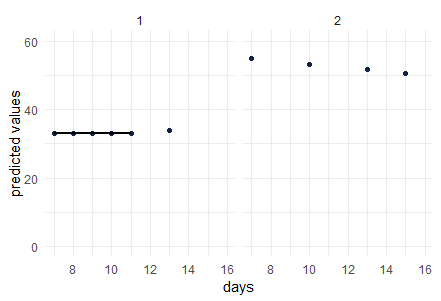
I created a multilevel regression model with nlme package and now I would like to illustrate the regression line obtained for some patients (unfortunately I cannot use geom_smooth with nlme).
So using the model I obtained the following predicted values (predicted_value) at different times (date_day) and here for two patients (ID1 and ID2).
df <- data.frame (ID = c (rep (1, 10), rep(2, 10)),
date_day = c (7:16, 7:16),
predicted_value = c (33, 33, 33, 33, 33, NA, 34, NA, NA, NA,
55, NA, NA, 53.3, NA, NA, 51.6, NA, 50.5, NA))
ID date_day predicted_value
1 1 7 33.0
2 1 8 33.0
3 1 9 33.0
4 1 10 33.0
5 1 11 33.0
6 1 12 NA
7 1 13 34.0
8 1 14 NA
9 1 15 NA
10 1 16 NA
11 2 7 55.0
12 2 8 NA
13 2 9 NA
14 2 10 53.3
15 2 11 NA
16 2 12 NA
17 2 13 51.6
18 2 14 NA
19 2 15 50.5
20 2 16 NA
Now I would like to draw the regression line for each of these patients. So I tried the following
ggplot(df%>% filter(ID %in% c("1", "2")))
aes(x = date_day, y = predicted_value)
geom_point(shape = "circle", size = 1.5, colour = "#112446", na.rm = T)
geom_line(aes(y = predicted_value), na.rm = T, size = 1)
theme_minimal()
facet_wrap(vars(ID))
scale_x_continuous(name="days", limits=c(7, 16))
scale_y_continuous(name="predicted values", limits=c(0, 60))
But I end with the following plots: patient 1 : the line is interrupted, and patient 2 no line at all. How can I fix that ?
Thanks a lot
CodePudding user response:
Thank you @BenBolker , indeed changing the first line
ggplot(df%>% filter(ID %in% c("1", "2")))
to
ggplot(na.omit(df)%>% filter(ID %in% c("1", "2")))
allowed to solve the job