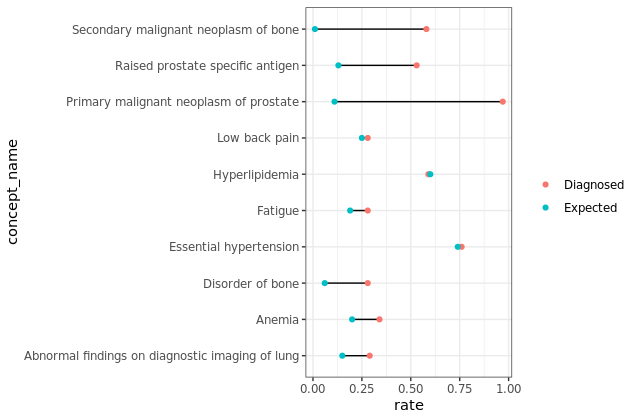
The below data has two groups diagnosed and Expected. I am creating a Cleveland dot plot to compare the diagnosed vs the expected value. I want to reorder the plot to show the concept_names based on the diagnosed rates descending which is how I have set the dataset.
concept_name rate grp
1 Disorder of bone 0.28 Diagnosed
2 Fatigue 0.28 Diagnosed
3 Low back pain 0.28 Diagnosed
4 Abnormal findings on diagnostic imaging of lung 0.29 Diagnosed
5 Anemia 0.34 Diagnosed
6 Raised prostate specific antigen 0.53 Diagnosed
7 Secondary malignant neoplasm of bone 0.58 Diagnosed
8 Hyperlipidemia 0.59 Diagnosed
9 Essential hypertension 0.76 Diagnosed
10 Primary malignant neoplasm of prostate 0.97 Diagnosed
11 Secondary malignant neoplasm of bone 0.01 Expected
12 Disorder of bone 0.06 Expected
13 Primary malignant neoplasm of prostate 0.11 Expected
14 Raised prostate specific antigen 0.13 Expected
15 Abnormal findings on diagnostic imaging of lung 0.15 Expected
16 Fatigue 0.19 Expected
17 Anemia 0.20 Expected
18 Low back pain 0.25 Expected
19 Hyperlipidemia 0.60 Expected
20 Essential hypertension 0.74 Expected
p <- ggplot(df, aes(rate, concept_name))
geom_line(aes(group = concept_name))
geom_point(aes(color = grp) )
theme_bw() theme (legend.title=element_blank())
The plot does not follow the order of the dataset. I would expect to see the concept names - Disorder of bone, Fatigue , Low back pain..
Data
structure(list(concept_name = structure(c(3L, 5L, 7L, 1L, 2L,
9L, 10L, 6L, 4L, 8L, 10L, 3L, 8L, 9L, 1L, 5L, 2L, 7L, 6L, 4L), .Label = c("Abnormal findings on diagnostic imaging of lung",
"Anemia", "Disorder of bone", "Essential hypertension", "Fatigue",
"Hyperlipidemia", "Low back pain", "Primary malignant neoplasm of prostate",
"Raised prostate specific antigen", "Secondary malignant neoplasm of bone"
), class = "factor", scores = structure(c(`Abnormal findings on diagnostic imaging of lung` = NA_real_,
Anemia = NA_real_, `Disorder of bone` = NA_real_, `Essential hypertension` = NA_real_,
Fatigue = NA_real_, Hyperlipidemia = NA_real_, `Low back pain` = NA_real_,
`Primary malignant neoplasm of prostate` = NA_real_, `Raised prostate specific antigen` = NA_real_,
`Secondary malignant neoplasm of bone` = NA_real_), .Dim = 10L, .Dimnames = list(
c("Abnormal findings on diagnostic imaging of lung", "Anemia",
"Disorder of bone", "Essential hypertension", "Fatigue",
"Hyperlipidemia", "Low back pain", "Primary malignant neoplasm of prostate",
"Raised prostate specific antigen", "Secondary malignant neoplasm of bone"
)))), rate = c(0.28, 0.28, 0.28, 0.29, 0.34, 0.53, 0.58,
0.59, 0.76, 0.97, 0.01, 0.06, 0.11, 0.13, 0.15, 0.19, 0.2, 0.25,
0.6, 0.74), grp = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L), .Label = c("Diagnosed",
"Expected"), class = "factor")), class = "data.frame", row.names = c(NA,
-20L))
CodePudding user response:
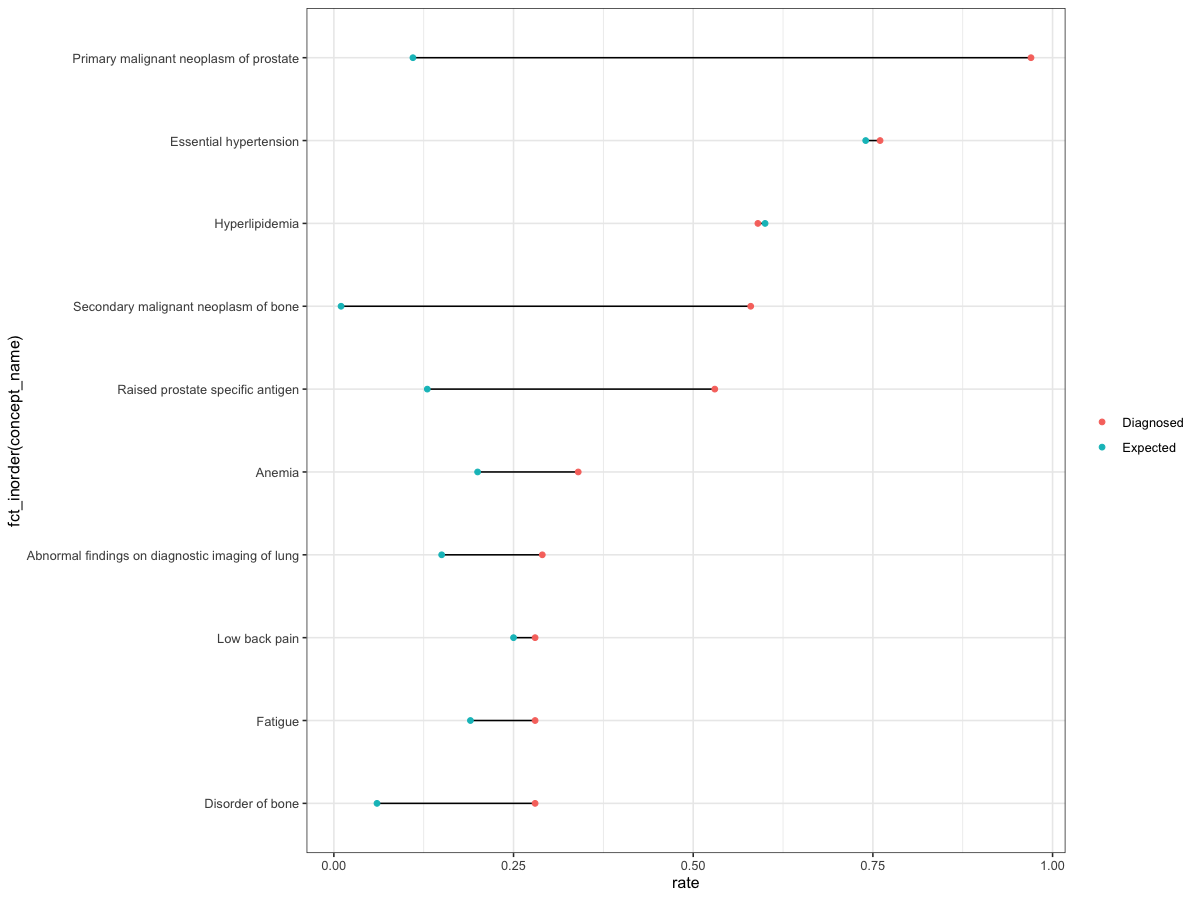
You can use fct_inorder from forcats:
library(tidyverse)
ggplot(df, aes(rate, fct_inorder(concept_name)))
geom_line(aes(group = concept_name))
geom_point(aes(color = grp))
theme_bw()
theme (legend.title = element_blank())
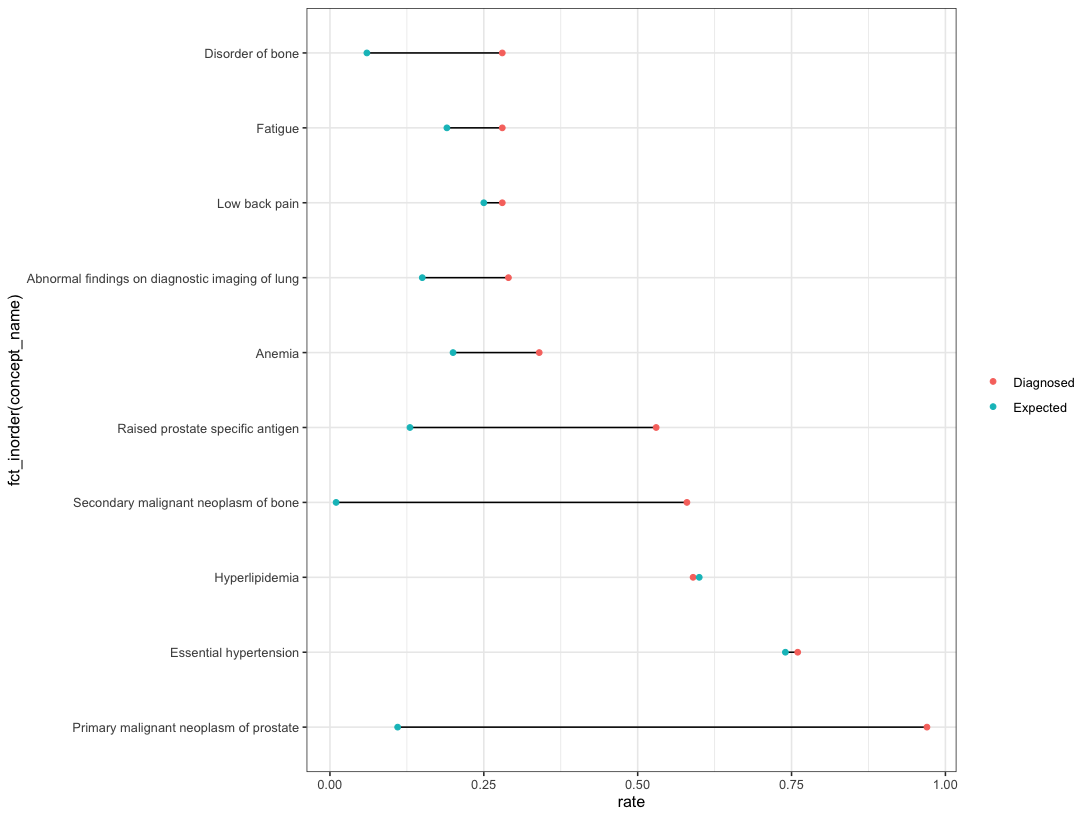
Or if you are wanting to have the order from the top of the y-axis, then we can first arrange the data, then use fct_inorder:
df %>%
arrange(grp, desc(rate), desc(concept_name)) %>%
ggplot(aes(rate, fct_inorder(concept_name)))
geom_line(aes(group = concept_name))
geom_point(aes(color = grp))
theme_bw()
theme (legend.title = element_blank())