Background
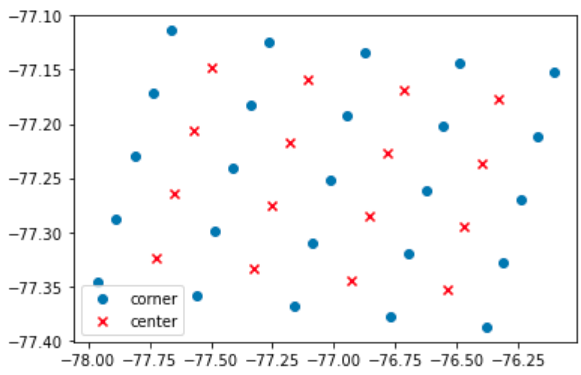
I have four 2D arrays: lon_centers, lat_centers, lon_bnds, and lat_bnds. bnds means the boundary of each pixel and centers stands for the center of pixels.
Data overview
import numpy as np
import matplotlib.pyplot as plt
lon_bnds = np.array([[-77.9645 , -77.56074 , -77.162025, -76.76827 , -76.37937 ],
[-77.88815 , -77.48613 , -77.08915 , -76.69711 , -76.30993 ],
[-77.811676, -77.41139 , -77.01614 , -76.62582 , -76.24034 ],
[-77.73638 , -77.337814, -76.944275, -76.55565 , -76.17186 ],
[-77.66197 , -77.265114, -76.87326 , -76.48632 , -76.1042 ]])
lat_bnds = np.array([[-77.34674 , -77.35804 , -77.36858 , -77.378395, -77.38752 ],
[-77.28847 , -77.299614, -77.31001 , -77.31969 , -77.328674],
[-77.23022 , -77.24122 , -77.25147 , -77.26101 , -77.26986 ],
[-77.17193 , -77.182785, -77.192894, -77.20229 , -77.211006],
[-77.11363 , -77.12434 , -77.13431 , -77.14357 , -77.15215 ]])
lon_centers = np.array([[-77.72404 , -77.323685, -76.92833 , -76.53787 ],
[-77.64892 , -77.2503 , -76.85666 , -76.46791 ],
[-77.57335 , -77.17646 , -76.78454 , -76.3975 ],
[-77.499626, -77.10444 , -76.71421 , -76.32886 ]])
lat_centers = np.array([[-77.32333 , -77.334175, -77.344284, -77.353676],
[-77.264946, -77.27564 , -77.28561 , -77.29487 ],
[-77.20665 , -77.21719 , -77.22702 , -77.23614 ],
[-77.14826 , -77.15867 , -77.16835 , -77.17734 ]])
plt.scatter(lon_bnds, lat_bnds, label='corner')
plt.scatter(lon_centers, lat_centers, marker='x', c='r', label='center')
plt.legend()
Goal
The goal is to calculate the area (m2) of each pixel, which means area has the same shape as lon_centers and lat_centers.
I found this useful 
A = 0.5 * ((x[1:, :-1] * y[:-1, :-1] x[1:, 1:] * y[1:, :-1] x[:-1, 1:] * y[1:, 1:] x[:-1, :-1] * y[:-1, 1:]) - (x[:-1, :-1] * y[1:, :-1] x[1:, :-1] * y[1:, 1:] x[1:, 1:] * y[:-1, 1:] x[:-1, 1:] * y[:-1, :-1]))
The result is
array([[65.80938398, 64.91795373, 64.04493782, 63.19815103],
[65.77719064, 64.87959934, 64.01155424, 63.16247495],
[65.77861439, 64.87956589, 64.01289216, 63.16872493],
[65.755155 , 64.85808632, 63.98664385, 63.14105705]])
Units are square kilometers.
For larger quadrangles, you may want to project each one individually about its center. In that case, you can do something like this:
def xy(s1, s2):
x = R[s1, s2] * (lons[s1, s2] - lon_c) * np.cos(lats[s1, s2])
y = R[s1, s2] * (lats[s1, s2] - lat_c)
return x, y
x1, y1 = xy(slice(None, -1), slice(None, -1))
x2, y2 = xy(slice(None, -1), slice(1, None))
x3, y3 = xy(slice(1, None), slice(1, None))
x4, y4 = xy(slice(1, None), slice(None, -1))
A = 0.5 * ((x1 * y2 x2 * y3 x3 * y4 x4 * y1) -
(x2 * y1 x3 * y2 x4 * y3 x1 * y4))
In this case, the result is similar:
array([[65.80950493, 64.9180908 , 64.04508934, 63.19831548],
[65.77721436, 64.87964039, 64.01161095, 63.16254575],
[65.77854026, 64.87951032, 64.01285345, 63.16870148],
[65.75498209, 64.85793324, 63.98650883, 63.14093847]])
The areas computed this way are almost identical:
array([[65.80950493, 64.9180908 , 64.04508934, 63.19831548],
[65.77721436, 64.87964039, 64.01161095, 63.16254575],
[65.77854026, 64.87951032, 64.01285345, 63.16870148],
[65.75498209, 64.85793324, 63.98650883, 63.14093847]])
The differences are just a few tens of square meters:
array([[-1.20941134e-04, -1.37064710e-04, -1.51528127e-04, -1.64456803e-04],
[-2.37217200e-05, -4.10480434e-05, -5.67132960e-05, -7.08008247e-05],
[ 7.41315753e-05, 5.55721762e-05, 3.87051454e-05, 2.34522307e-05],
[ 1.72913145e-04, 1.53083044e-04, 1.35020126e-04, 1.18583183e-04]])
CodePudding user response:
Here's something you can consider when you are doing multiple for loops onto arrays.
# Without caching
def function1():
geod = Geod(ellps="WGS84")
areas = []
for x in range(lon_bnds.shape[0]-1):
for y in range(lon_bnds.shape[1]-1):
lons = lon_bnds[x:x 2, y:y 2].ravel()
lats = lat_bnds[x:x 2, y:y 2].ravel()
lons[-2], lons[-1] = lons[-1], lons[-2]
lats[-2], lats[-1] = lats[-1], lats[-2]
poly_area, poly_perimeter = geod.polygon_area_perimeter(lons, lats)
areas.append(poly_area)
return areas
# With caching
from functools import lru_cache
@lru_cache(maxsize=512)
def function2():
geod = Geod(ellps="WGS84")
areas = []
for x in range(lon_bnds.shape[0]-1):
for y in range(lon_bnds.shape[1]-1):
lons = lon_bnds[x:x 2, y:y 2].ravel()
lats = lat_bnds[x:x 2, y:y 2].ravel()
lons[-2], lons[-1] = lons[-1], lons[-2]
lats[-2], lats[-1] = lats[-1], lats[-2]
poly_area, poly_perimeter = geod.polygon_area_perimeter(lons, lats)
areas.append(poly_area)
return areas
%timeit function1()
229 µs ± 10.8 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
%timeit function2()
84.3 ns ± 2.8 ns per loop (mean ± std. dev. of 7 runs, 10000000 loops each)