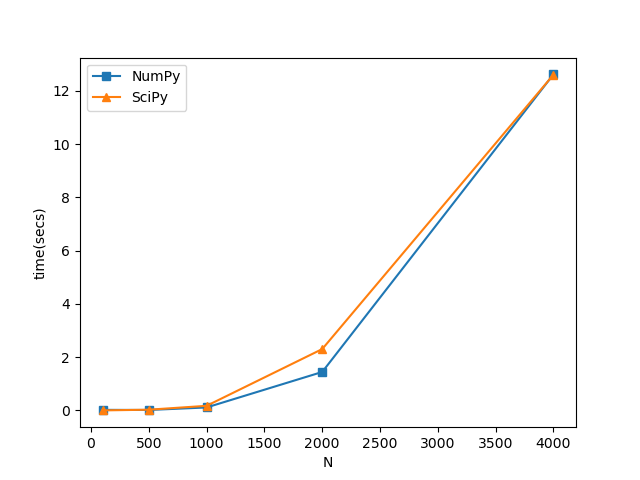
I have to make use of diagonalization routines to obtain all eigenpairs of a Hermitian complex matrix. I am a bit limited by performance since I need to repeat the operation thousands of times and my matrices are roughly of 8000x8000. I have created a little comparison between the NumPy and SciPy routines for diagonalization of Hermitian matrices and I got these times on a 6 physical cores machine:
I am observing that for 8000x8000 matrices this scales to~0.8 minutes and I need to repeat the process 50000 times. Is there something I am missing here or are these actually the performance times? Overall, this all looks very slow specially if this needs to be repeated several times. In fact, on a 30 core machine, I observe little performance gain. I am using the Python3.8 under Anaconda distribution so this is linked against the MKL.
Here is the example code
import numpy as np
import scipy.linalg
import matplotlib.pyplot as pyt
from time import time
t_ls = []
d_ls = np.array([100, 500, 1000,2000,4000])
for N in d_ls:
A =np.random.rand( N,N ) 1j*np.random.rand( N,N )
A = 0.5*( A np.conj(A.T) )
ts = time()
evals, evecs = np.linalg.eigh( A )
t_np = time()-ts
ts = time()
evals2, evecs2 = scipy.linalg.eigh( A )
t_sp = time()-ts
t_ls.append(np.array([t_np, t_sp]))
t_ls = np.array(t_ls)
pyt.plot( d_ls, t_ls[:,0], marker='s' )
pyt.plot( d_ls, t_ls[:,1], marker='^')
pyt.xlabel("N")
pyt.ylabel("time(secs)")
pyt.legend(["NumPy", "SciPy"])
pyt.show()
USING SVD AND MP PARALELLIZATION
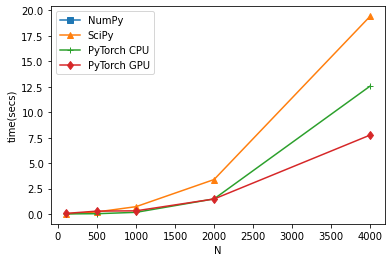
Going through some of the comments in the post, I have tried SVD of the matrix and multiprocessing. I all cases I still see the serialized approach with NumPy eigh is the most efficient one; here is the code:
import numpy as np
import scipy.linalg
import matplotlib.pyplot as pyt
from time import time
import psutil
def f_mp_pool(*args):
N = args[0]
A =np.random.rand( N,N ) 1j*np.random.rand( N,N )
A = 0.5*( A np.conj(A.T) )
evals, evecs = np.linalg.eigh(A)
return evals, evecs
nreps = 100
N = 700
ts = time()
for n in range(nreps):
A =np.random.rand( N,N ) 1j*np.random.rand( N,N )
A = 0.5*( A np.conj(A.T) )
res = np.linalg.eigh(A)
print("serialized:", time()-ts)
#use svd
import scipy.linalg
ts = time()
for n in range(nreps):
res = scipy.linalg.svd( A, full_matrices=True, check_finite=False )
print("SVD:", time()-ts)
import multiprocessing as mp
nproc = psutil.cpu_count(logical=False)-1
mp_pool = mp.Pool(processes=nproc)
args_ls = [ (N,) for n in range(nreps) ]
ts = time()
res = mp_pool.starmap( f_mp_pool, args_ls )
print("parallel:", time()-ts)
CodePudding user response: