Hey people of the web,
I have a function i'm creating 'show_monthly_temp', in which i'm attempting to plot a figure with 12 scatter sub plots, where each subplot's purpose mentioned in the next lines.
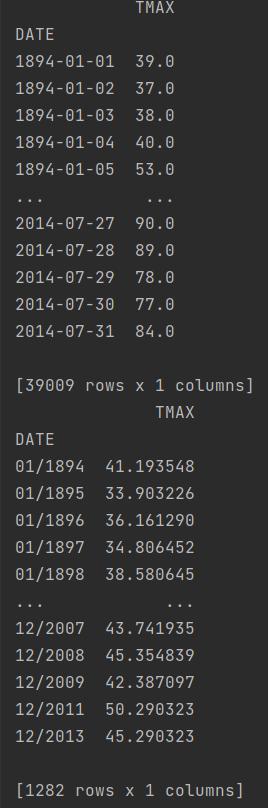
which prints the following thing (DATE is a datetime object):
Now, what I attempt to do (unsuccessfully) is to plot 12 subplots representing the average of each month of the year throughout the years of sampling.
Any ideas of how I could do so?
Just to emphasise what i'm aiming for:
each subplot is a month, where Y will represent the temperature during that year of measurement, and X the year of the sample.
Thanks! :-)
full code is here:
import pandas as pd
from datetime import datetime
import numpy as np
import scipy as stats
import matplotlib.pyplot as plt
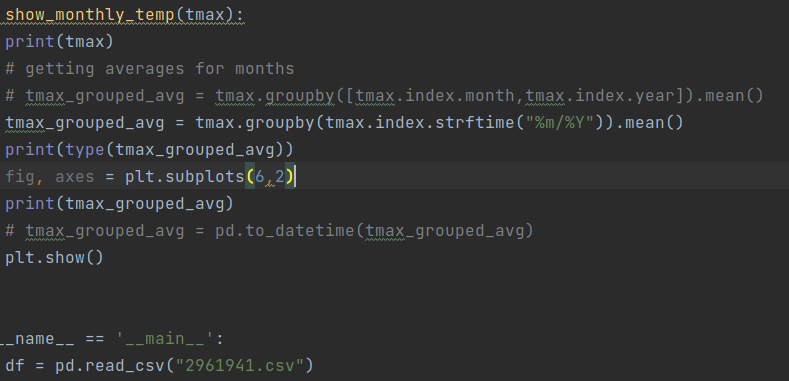
def show_monthly_temp(tmax):
print(tmax)
# getting averages for months
# tmax_grouped_avg = tmax.groupby([tmax.index.month,tmax.index.year]).mean()
tmax_grouped_avg = tmax.groupby(tmax.index.strftime("%m/%Y")).mean()
print(type(tmax_grouped_avg))
fig, axes = plt.subplots(6,2)
print(tmax_grouped_avg)
# tmax_grouped_avg = pd.to_datetime(tmax_grouped_avg)
plt.show()
if __name__ == '__main__':
df = pd.read_csv("2961941.csv")
# set date column as index, drop the 'DATE' column to avoid repititions create as datetime object
# speed up parsing using infer_datetime_format=True.
df.index = pd.to_datetime(df['DATE'], infer_datetime_format=True)
# create new tables
tmax = df.filter(['TMAX'], axis=1).dropna()
snow = df.filter(['SNOW']).dropna()
# count number of snow day samples - make sure at least >= 28
snow_grouped = snow.groupby(pd.Grouper(level='DATE', freq="M")).transform('count')
snow = (snow[snow_grouped['SNOW'] >= 28])
# count number of tmax day samples - make sure at least >= 28
tmax_grouped = tmax.groupby(pd.Grouper(level='DATE', freq="M")).transform('count')
tmax = (tmax[tmax_grouped['TMAX'] >= 28])
################ Until here - initialized data ###############
show_monthly_temp(tmax)
CodePudding user response:
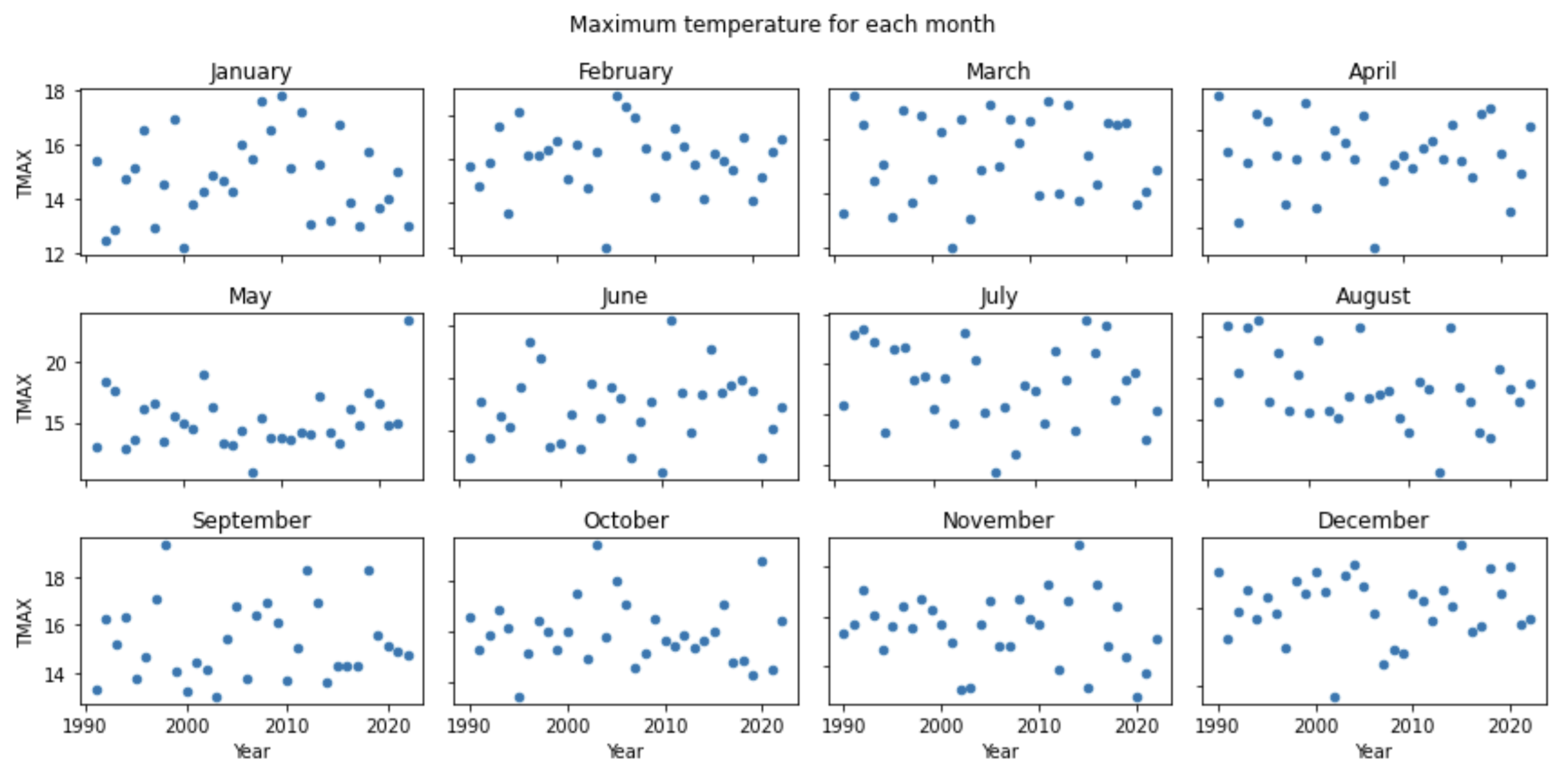
If I understood correctly, you could do it like this:
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import calendar
# Fake data - generating 'tmax_grouped_avg'.
df = pd.DataFrame({'Date': pd.date_range('1990-10-01', '2023-05-01')})
df['TMAX'] = np.random.random((len(df)))*30
tmax_grouped_avg = df.groupby(df['Date'].dt.strftime('%m/%Y')).mean()
# New code here.
tmax_grouped_avg['datetime'] = pd.to_datetime(tmax_grouped_avg.index)
tmax_grouped_avg['Year'] = tmax_grouped_avg['datetime'].dt.year
groups = tmax_grouped_avg.groupby(tmax_grouped_avg['datetime'].dt.month)
f, axes = plt.subplots(nrows=3, ncols=4, figsize=(12, 6))
for (grp_id, grp_df), ax in zip(groups, axes.ravel()):
# Check if you want sharex and sharey.
grp_df.plot.scatter(ax=ax, x='Year', y='TMAX', title=f'{calendar.month_name[grp_id]}', legend=False,
sharey=True, sharex=True)
plt.suptitle('Maximum temperature for each month')
plt.tight_layout()
plt.show()