I want to plot the data shown below as a grouped bar_plot.
I tried position = "dodge" or position = "dodge2" but it didn't work. Ι also tried position = position_dodge()
It kinda works if i use geom_bar instead of geom_col and remove the y=overlap_percent:
p3 <- ggplot(data = comp_coors, aes(x = species_code, fill = mirna_form))
geom_bar(position = "dodge2") theme_classic()
p3
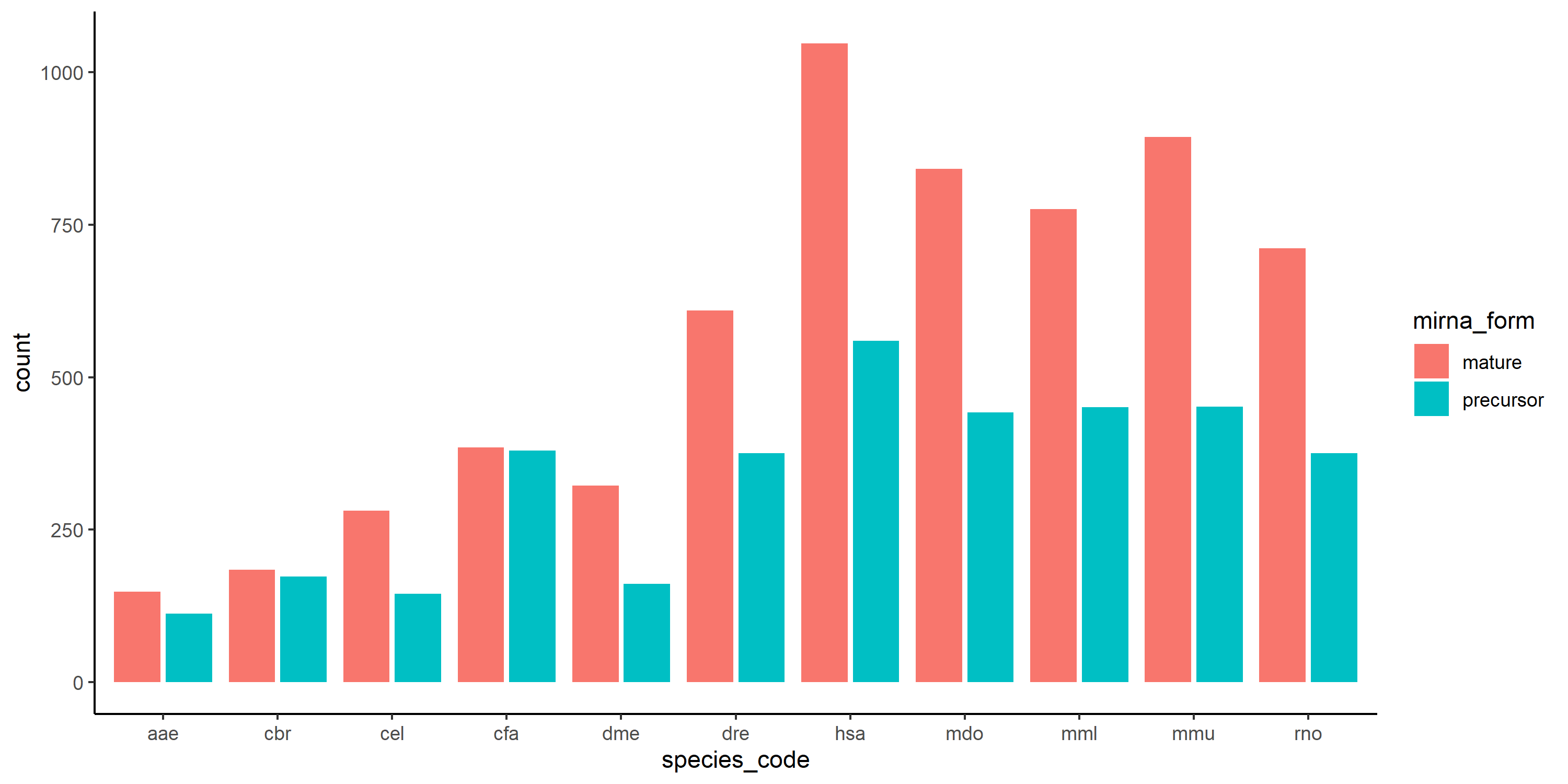
but i would like the y_axis to have the overlap_percent.
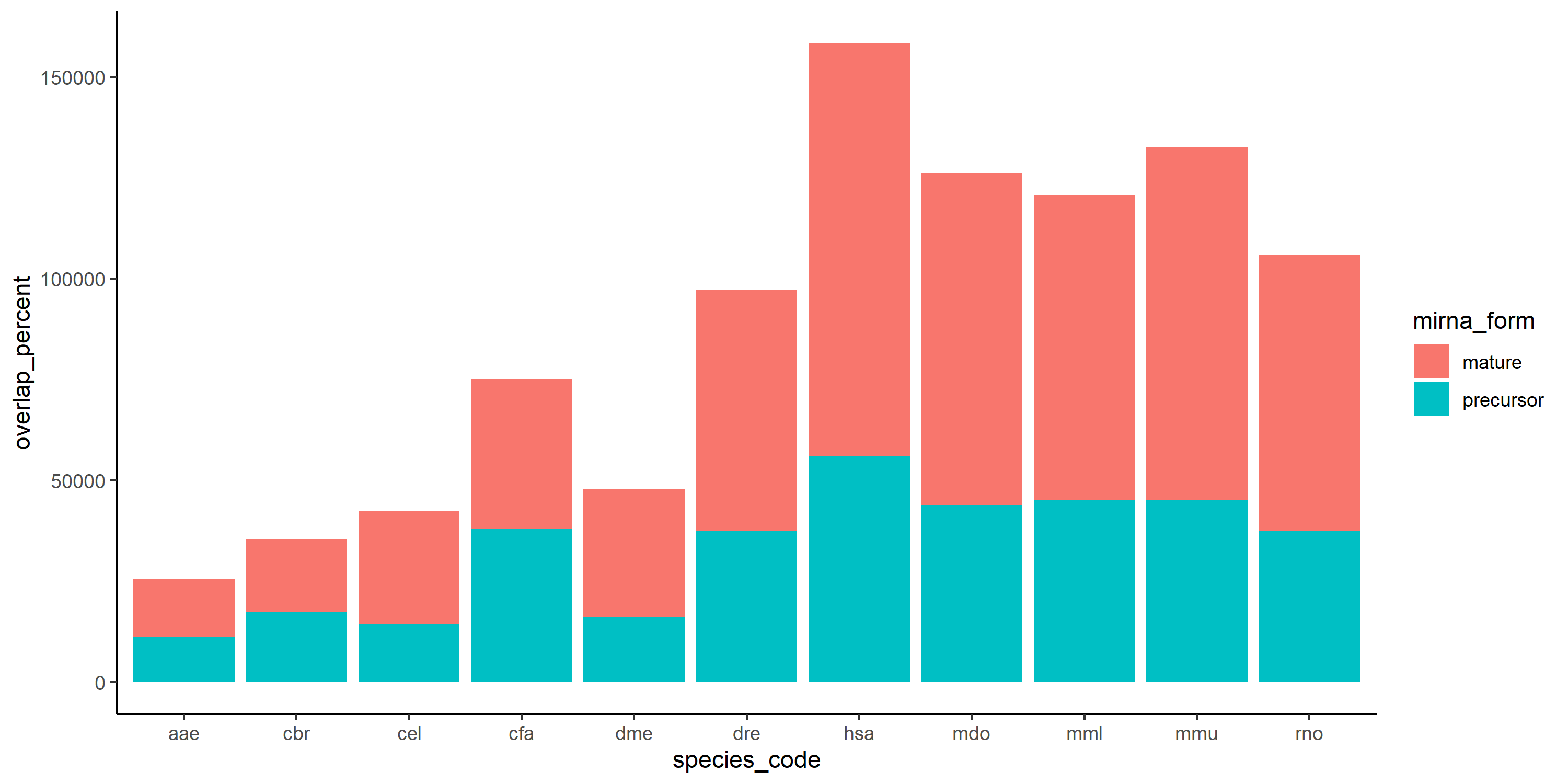
Another attempt which ends in a stacked barplot is:
p2 <- ggplot(data = comp_coors, aes(x = species_code, y = overlap_percent, fill = mirna_form))
geom_bar(stat = "identity") theme_classic()
p2
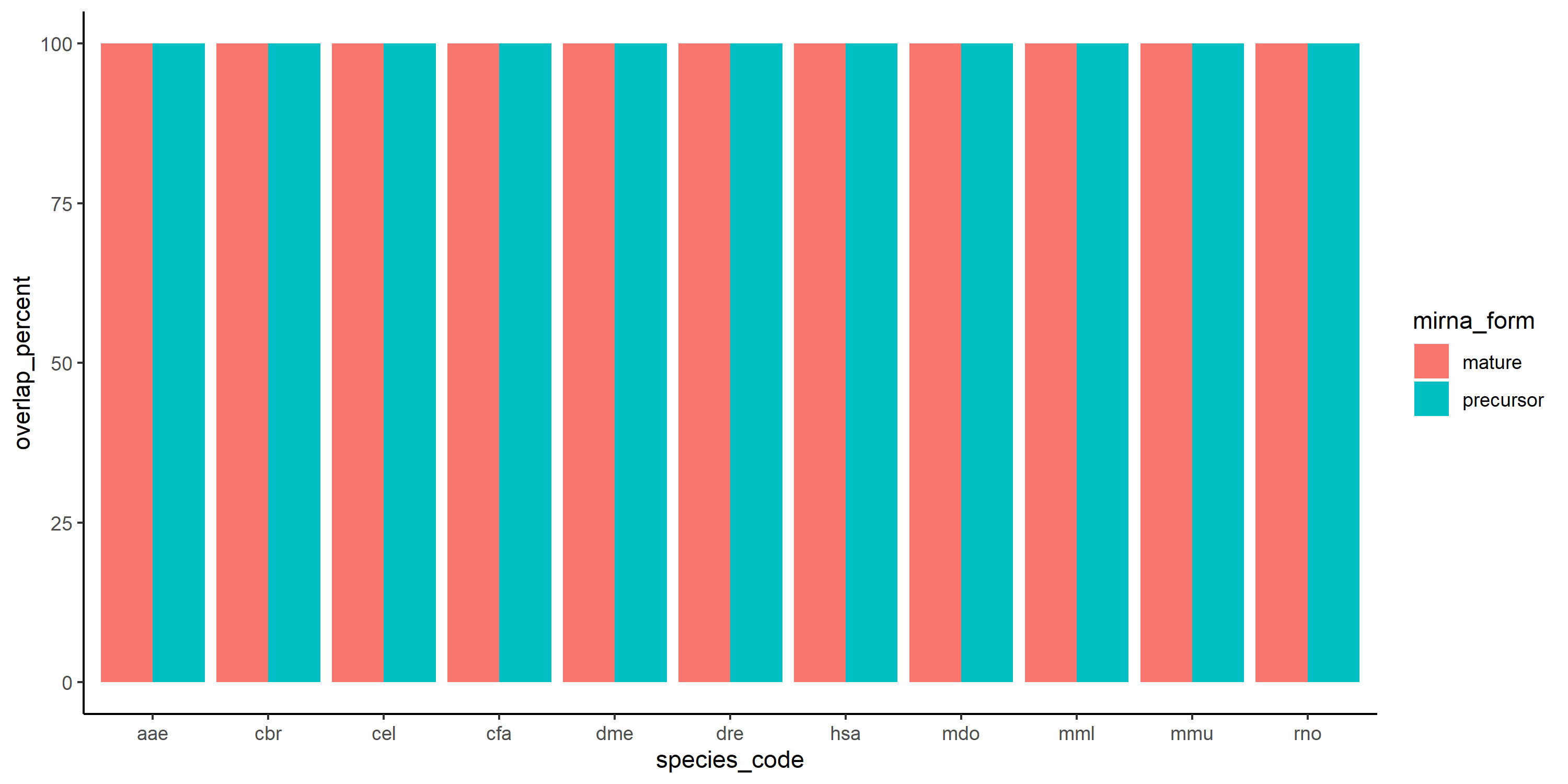
Finally by using geom_col, it returns this which it doesn't make sense, at least to me:
p4 <- ggplot(data = comp_coors, aes(x = species_code, y = overlap_percent, fill = mirna_form))
geom_col(position = "dodge") theme_classic()
p4
The data that i want to plot :
comp_coors <- data.table( species = c("aae","cel", "dme","hsa", "mdo"),
mirna_form = c("mature", "precursor"),
overlap_percent = c(100.0, 100.0, 88.0, 95.5, 91.7, 100.0, 96.6, 98.4),
overlapping_attribute = c("ID=MIMAT0014285;Alias=MIMAT0014285", "ID=MI0000043;Alias=MI0000043;Name=cel-mir-72", "ID=MIMAT0000401;Alias=MIMAT0000401;Name=dme-miR-", "ID=MI0000791;Alias=MI0000791;Name=hsa-mir-383", "ID=MI0005331;Alias=MI0005331;Name=mdo-let-7g")
)
CodePudding user response:
Try using species as a factor and add stat = "identity" like this:
ggplot(data = comp_coors, aes(x = factor(species), y = overlap_percent, fill = mirna_form))
geom_bar(position = "dodge", stat = "identity") theme_classic() labs(x = "Species", y = "Overlap percent")
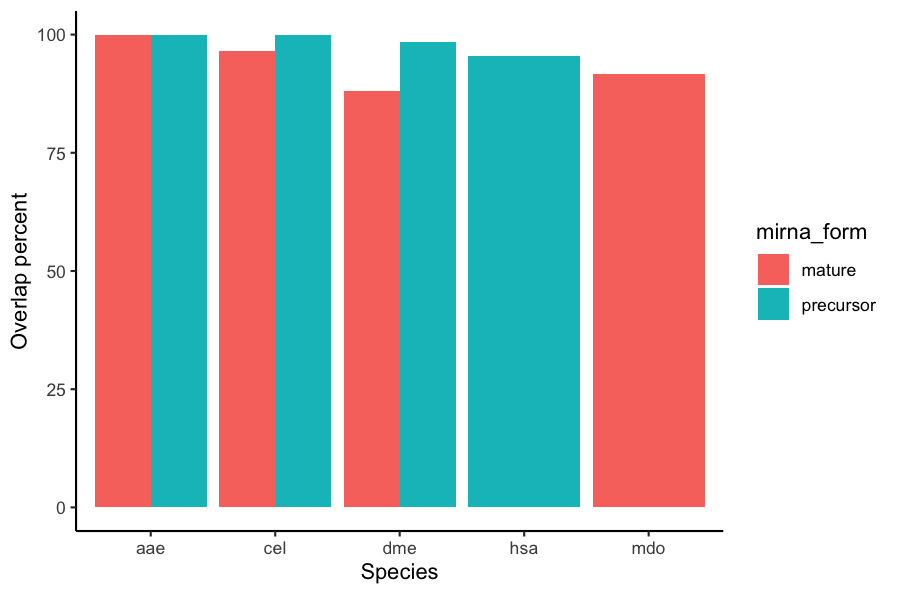
Output:
A grouped barplot with overlap_percent on y-axis right.