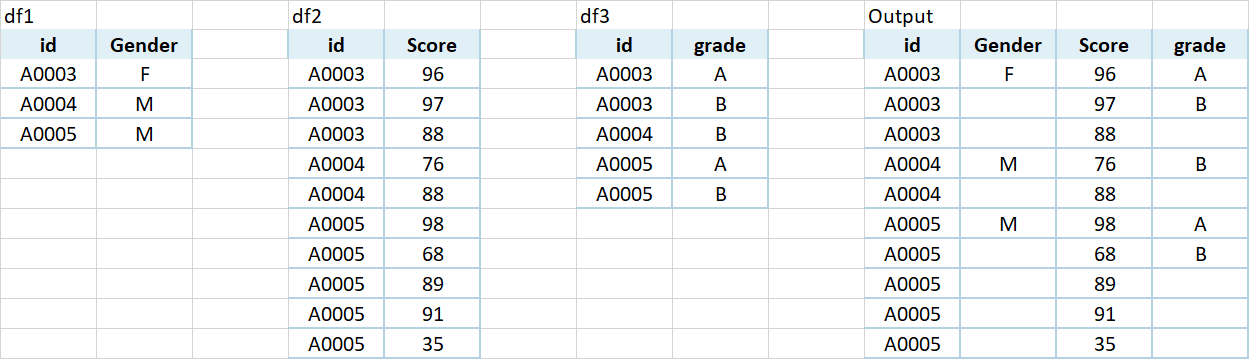
I have 3 dfs with different length. I would like to bind thme together by id. there is no row connection among those 3 dfs. I did not have a clue how to do it. Could anyone guide me on this? Thanks.
df1<-structure(list(id = c("A0003", "A0004", "A0005"), Gender = c("F",
"M", "M")), row.names = c(NA, -3L), class = c("tbl_df", "tbl",
"data.frame"))
df2<-structure(list(id = c("A0003", "A0003", "A0003", "A0004", "A0004",
"A0005", "A0005", "A0005", "A0005", "A0005"), Score = c(96, 97,
88, 76, 88, 98, 68, 89, 91, 35)), row.names = c(NA, -10L), class = c("tbl_df",
"tbl", "data.frame"))
df3<-structure(list(id = c("A0003", "A0003", "A0004", "A0005", "A0005",
"A0005"), grade = c("A", "B", "B", "A", "B", "F")), row.names = c(NA,
-6L), class = c("tbl_df", "tbl", "data.frame"))
CodePudding user response:
We may need to create a sequence column in each dataset by id as there are duplicates for 'id' in 'df2' and 'df3'. Keep all the datasets in a list, then loop over the list with map (from purrr), create a row sequence (rowid from data.table), reduce the list elements to a single dataset by joining (full_join) by 'id' and the row id ('rid')
library(dplyr)
library(purrr)
library(data.table)
lst(df1, df2, df3) %>%
map(~.x %>%
mutate(rid = rowid(id))) %>%
reduce(full_join, by = c('id', 'rid')) %>%
select(-rid) %>%
arrange(id)
-output
# A tibble: 10 × 4
id Gender Score grade
<chr> <chr> <dbl> <chr>
1 A0003 F 96 A
2 A0003 <NA> 97 B
3 A0003 <NA> 88 <NA>
4 A0004 M 76 B
5 A0004 <NA> 88 <NA>
6 A0005 M 98 A
7 A0005 <NA> 68 B
8 A0005 <NA> 89 F
9 A0005 <NA> 91 <NA>
10 A0005 <NA> 35 <NA>
CodePudding user response:
Another possible solution, based on dplyr:
library(dplyr)
bind_rows(df1, df2, df3) %>%
group_by(id) %>%
mutate(Score = lead(Score, 1)) %>%
mutate(grade = lead(grade, n_distinct(data.frame(id, Gender, Score)))) %>%
arrange(id) %>%
filter(!if_all(everything(), is.na)) %>%
ungroup
#> # A tibble: 10 × 4
#> id Gender Score grade
#> <chr> <chr> <dbl> <chr>
#> 1 A0003 F 96 A
#> 2 A0003 <NA> 97 B
#> 3 A0003 <NA> 88 <NA>
#> 4 A0004 M 76 B
#> 5 A0004 <NA> 88 <NA>
#> 6 A0005 M 98 A
#> 7 A0005 <NA> 68 B
#> 8 A0005 <NA> 89 F
#> 9 A0005 <NA> 91 <NA>
#> 10 A0005 <NA> 35 <NA>