I've looked up and down but can't find an example that closely resembles what I'm trying to do. In short, I have multiple trials of an experiment with multiple variables. The data is collected and stored in a long dataframe object, like so (generated data example):
Input:
import pandas as pd
import itertools
from scipy import rand
trials = ['trial_1' , 'trial_2']
ppi = ['5_ppi', '10_ppi']
rd = ['7%', '12%']
iter = [rd, ppi, trials]
df = pd.DataFrame()
for k, params in enumerate(list(itertools.product(*iter))):
ext_data = [rand(), rand()]
load_data = [rand(), rand()]
df1 = pd.DataFrame({'ext': ext_data, 'load': load_data, 'trial': params[2], 'ppi': params[1], 'rd': params[0]})
df = pd.concat([df, df1], axis=0)
print(df)
Output:
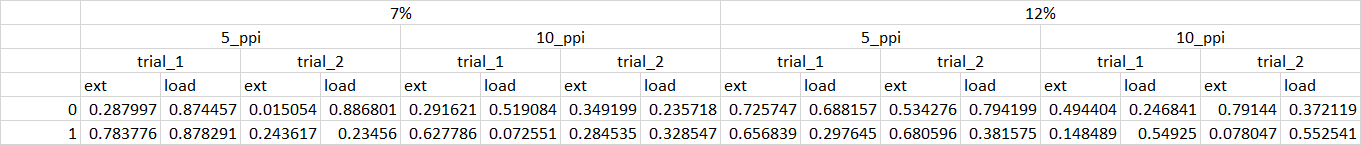
df
ext load trial ppi rd
0 0.287997 0.874457 trial_1 5_ppi 7%
1 0.783776 0.878291 trial_1 5_ppi 7%
0 0.015054 0.886801 trial_2 5_ppi 7%
1 0.243617 0.234560 trial_2 5_ppi 7%
0 0.291621 0.519084 trial_1 10_ppi 7%
1 0.627786 0.072551 trial_1 10_ppi 7%
0 0.349199 0.235718 trial_2 10_ppi 7%
1 0.284535 0.328547 trial_2 10_ppi 7%
0 0.725747 0.688157 trial_1 5_ppi 12%
1 0.656839 0.297645 trial_1 5_ppi 12%
0 0.534276 0.794199 trial_2 5_ppi 12%
1 0.680596 0.381575 trial_2 5_ppi 12%
0 0.494404 0.246841 trial_1 10_ppi 12%
1 0.148489 0.549250 trial_1 10_ppi 12%
0 0.791440 0.372119 trial_2 10_ppi 12%
1 0.078047 0.552541 trial_2 10_ppi 12%
[16 rows x 5 columns]
(in reality there are hundreds of data points for each trial, not just two like shown here)
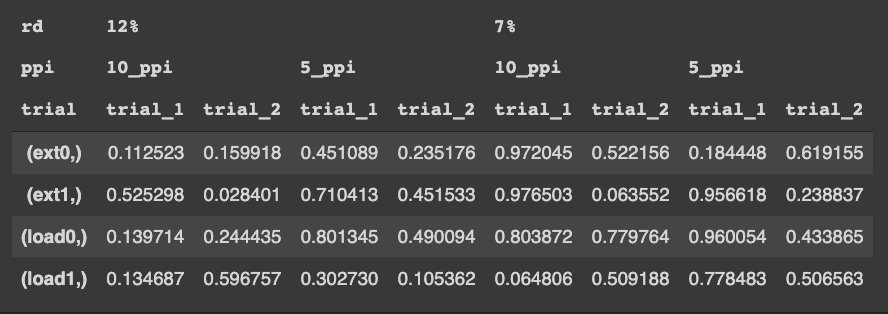
I would like to convert this to a wide dataframe with hierarchical columns, organized like so (i can't use markdown to make values/headers span multiple columns, so please forgive the screenshot)
I've tried so many things that I've lost track. I think pivot_table got me the closest to what I want, but it winds up aggregating the mean of the values instead of listing them out, like so:
Input:
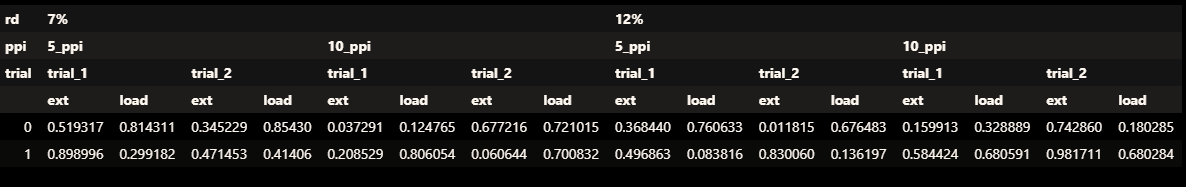
df.pivot_table(columns=['rd', 'ppi', 'trial'])
Output:
rd 12% 7%
ppi 10_ppi 5_ppi 10_ppi 5_ppi
trial trial_1 trial_2 trial_1 trial_2 trial_1 trial_2 trial_1 trial_2
ext 0.619217 0.661812 0.652555 0.167241 0.340024 0.42324 0.565166 0.436858
load 0.397430 0.102965 0.528162 0.550871 0.206560 0.28524 0.731204 0.303079
[2 rows x 8 columns]
It is also possible that each set of data has a different number of data points (i.e. Trial 1 might contain 2 data points, while Trial 2 might have 3). I'm at my wits end. What is the magic command to turn this long dataframe object into a wide dataframe object with multiple hierarchical columns?
CodePudding user response:
This may not be the most efficient answer, but I think it is achieving your desired outcome and should work for any number of observations.
def reformat(df, column_name):
groups = df.groupby(['rd','ppi','trial'])[column_name].apply(list)
temp_1 = groups.reset_index(name = 'listvalues')
# Make names for each of your new columns
col_names = [column_name str(i) for i in np.arange(len(temp_1['listvalues'][0]))]
# Split listvalues into df where every item is its own column
listvalues = pd.DataFrame(temp_1["listvalues"].to_list(), columns=[col_names])
# Merge listvalues with the temp_1 df and get rid of the extra listvalues column
df = temp_1.join(listvalues)
del df['listvalues']
df = df.pivot_table(columns=['rd', 'ppi', 'trial'])
return df
# Do this twice and append to get the desired dataframe
df2 = reformat(df, 'ext')
df3 = reformat(df, 'load')
finaldf = df2.append(df3)
Where finaldf looks like:
CodePudding user response:
try this:
idx_cols = [*df][-1:-4:-1]
res = df.set_index(idx_cols [df.groupby(idx_cols).cumcount()])
res = res.stack().unstack([0,1,2,-1])
print(res)