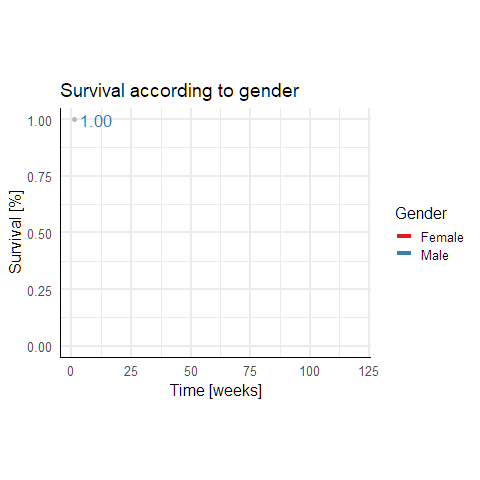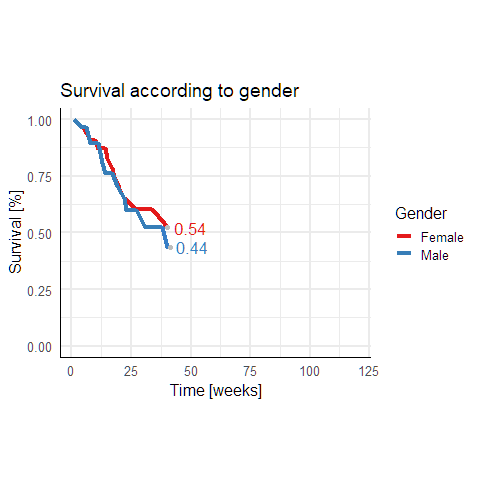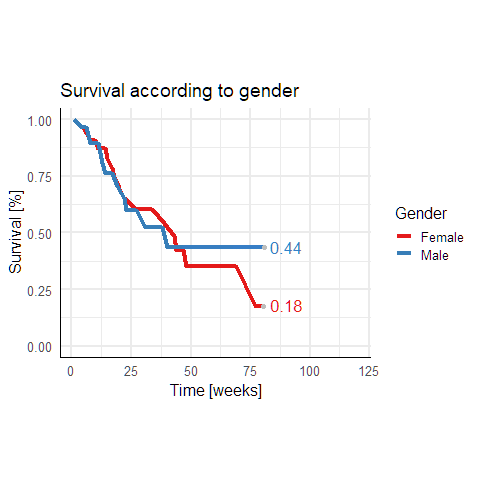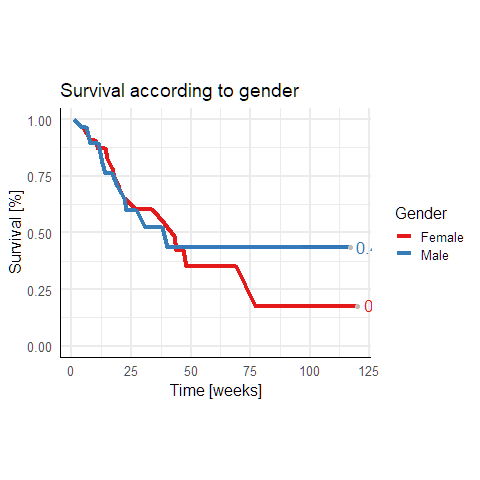
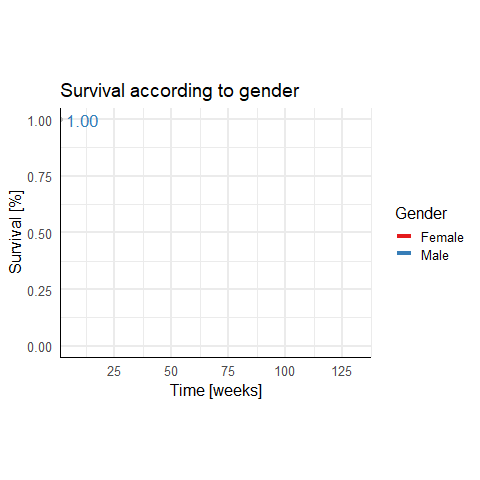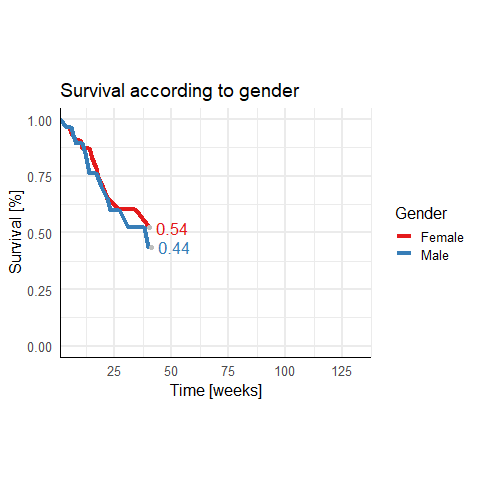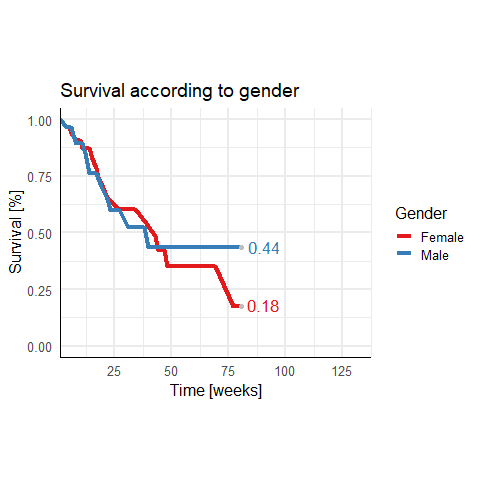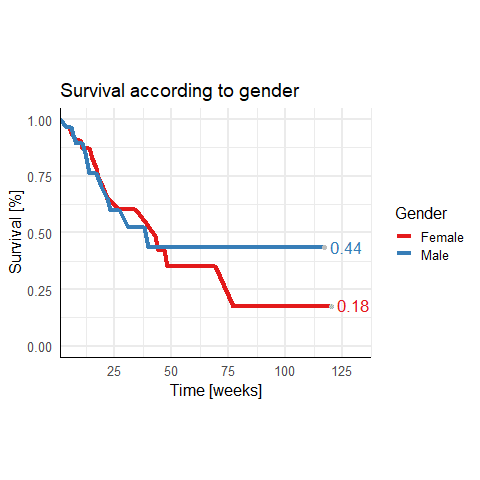
I have this survival dataframe:
df <- structure(list(time = c(5, 6, 8, 10, 11, 12, 13, 14, 15, 17,
18, 20, 22, 27, 31, 32, 34, 39, 43, 44, 47, 48, 50, 60, 69, 77,
120, 1, 4, 6, 7, 8, 9, 11, 12, 13, 14, 15, 17, 19, 22, 23, 25,
26, 27, 31, 38, 40, 64, 89, 96, 105, 117), surv = c(0.96969696969697,
0.93841642228739, 0.906057235311963, 0.906057235311963, 0.871208880107657,
0.871208880107657, 0.871208880107657, 0.871208880107657, 0.829722742959673,
0.786053124909164, 0.742383506858655, 0.695984537679989, 0.649585568501323,
0.603186599322657, 0.603186599322657, 0.603186599322657, 0.603186599322657,
0.542867939390391, 0.482549279458125, 0.42223061952586, 0.42223061952586,
0.351858849604883, 0.351858849604883, 0.351858849604883, 0.351858849604883,
0.175929424802442, 0.175929424802442, 1, 0.96551724137931, 0.96551724137931,
0.929757343550447, 0.892567049808429, 0.892567049808429, 0.892567049808429,
0.850063856960409, 0.805323653962492, 0.760583450964576, 0.760583450964576,
0.760583450964576, 0.706256061609964, 0.651928672255351, 0.597601282900738,
0.597601282900738, 0.597601282900738, 0.597601282900738, 0.522901122538146,
0.522901122538146, 0.435750935448455, 0.435750935448455, 0.435750935448455,
0.435750935448455, 0.435750935448455, 0.435750935448455), V3 = c(0.97,
0.94, 0.91, 0.91, 0.87, 0.87, 0.87, 0.87, 0.83, 0.79, 0.74, 0.7,
0.65, 0.6, 0.6, 0.6, 0.6, 0.54, 0.48, 0.42, 0.42, 0.35, 0.35,
0.35, 0.35, 0.18, 0.18, 1, 0.97, 0.97, 0.93, 0.89, 0.89, 0.89,
0.85, 0.81, 0.76, 0.76, 0.76, 0.71, 0.65, 0.6, 0.6, 0.6, 0.6,
0.52, 0.52, 0.44, 0.44, 0.44, 0.44, 0.44, 0.44), grupo = c(1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2), risk = c(33, 31, 29, 27, 26, 24,
23, 22, 21, 19, 18, 16, 15, 14, 13, 12, 11, 10, 9, 8, 7, 6, 5,
4, 3, 2, 1, 30, 29, 28, 27, 25, 24, 23, 21, 19, 18, 16, 15, 14,
13, 12, 11, 10, 9, 8, 7, 6, 5, 4, 3, 2, 1)), class = "data.frame", row.names = c(NA,
-53L))
With this code I constructed an animated Kaplan Meier plot:
library(ggplot2)
library(gganimate)
ggplot(data=df, aes(x=time, y=surv, group=grupo, color=as.factor(grupo)))
geom_line(size=1.5)
geom_point(size = 2, color="gray")
guides(color = guide_legend(
override.aes=list(shape = 1)))
scale_color_brewer(name="Gender",palette="Set1",labels=c("Female","Male"))
labs(title = "Survival according to gender", x = "Time [weeks]", y = "Survival [%]")
scale_y_continuous(limits = c(0,1))
geom_text(mapping=aes(y=V3,label=format(V3)),size=6,hjust=-0.2,vjust=0.5, show.legend = FALSE)
transition_reveal(time)
theme_minimal(base_size = 16)
theme(axis.line = element_line(colour = "black", size = 0.24),
aspect.ratio=4/5)
However at the end x=125 the numbers disappear. I think the grid is to short.
How can I expand the grid to avoid hiding the survival curve numbers at x = 125.
I have tried a few things like expanding x but could not solve it! I also changed hjust and vjust but I want to keep the numbers at this position!
CodePudding user response:
Instead of switching off clipping you can also expand the right upper limit of the x axis using expansion.
ggplot(df, aes(x = time, y = surv, group = grupo, color = as.factor(grupo)))
geom_line(size = 1.5)
geom_point(size = 2, color = "gray")
guides(color = guide_legend(override.aes = list(shape = 1)))
scale_color_brewer(
name = "Gender",palette = "Set1", labels = c("Female", "Male"))
labs(
title = "Survival according to gender",
x = "Time [weeks]",
y = "Survival [%]")
scale_y_continuous(limits = c(0, 1))
scale_x_continuous( # This is new
breaks = scales::breaks_width(25), # Label every 25 weeks
expand = expansion(mult = c(0, 0.15))) # 15% increase on the right
geom_text(
aes(y = V3, label = format(V3)),
size = 6,
hjust = -0.2,
vjust = 0.5,
show.legend = FALSE)
transition_reveal(time)
theme_minimal(base_size = 16)
theme(
axis.line = element_line(colour = "black", size = 0.24),
aspect.ratio = 4/5)
This increases the right upper limit by 15%.