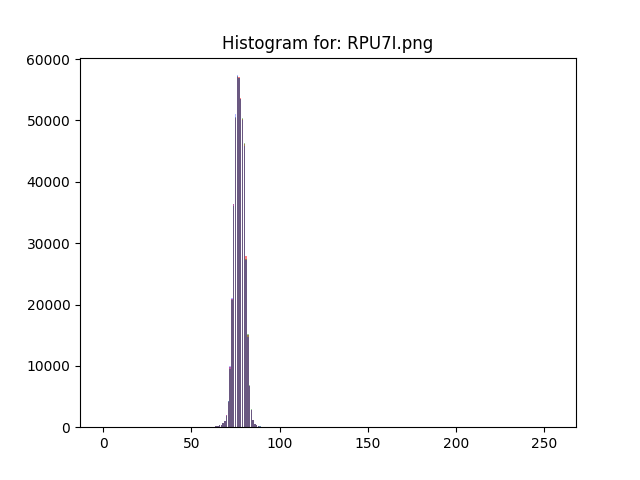
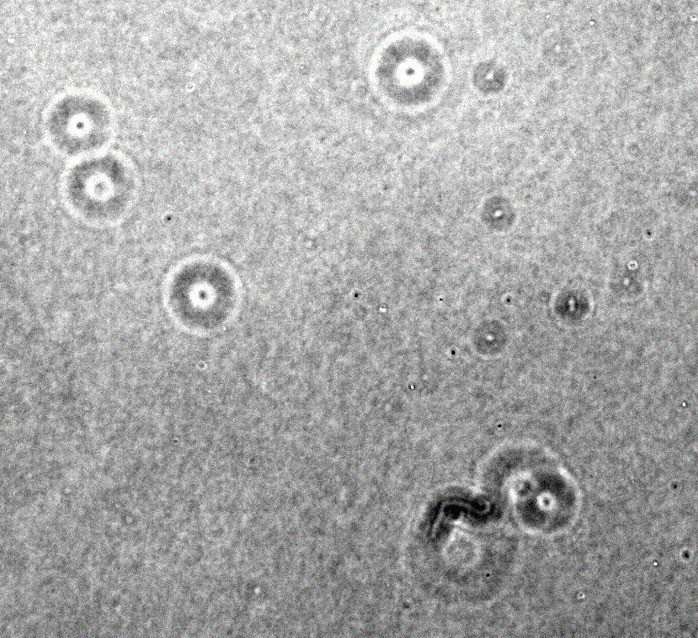
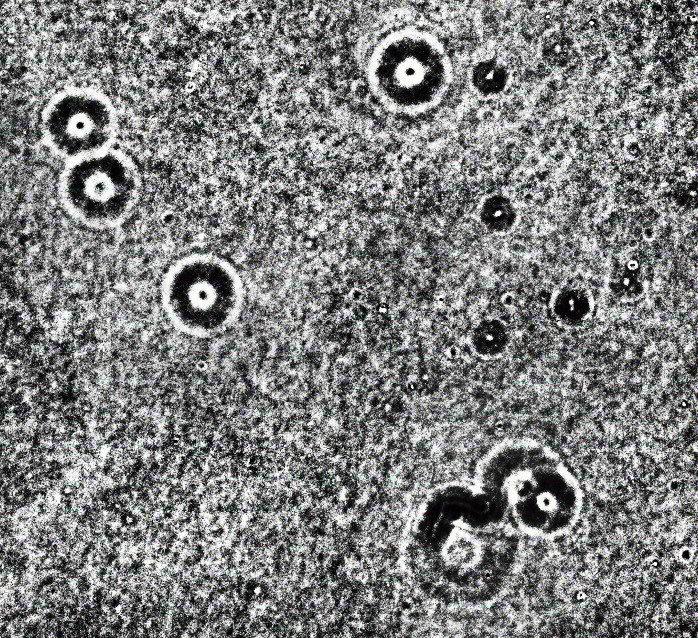
You clearly have a low-contrast image with all brightnesses in the range around 64-88, i.e. 25% to 33% of the 0..255 range. So my first instinct is to stretch the contrast so that 64 becomes zero and 88 becomes 255. I am doing it with ImageMagick here because it is conceptual and you/we can do it with OpenCV later if necessary.
magick RPU7I.png -level 25,33% 1.jpg
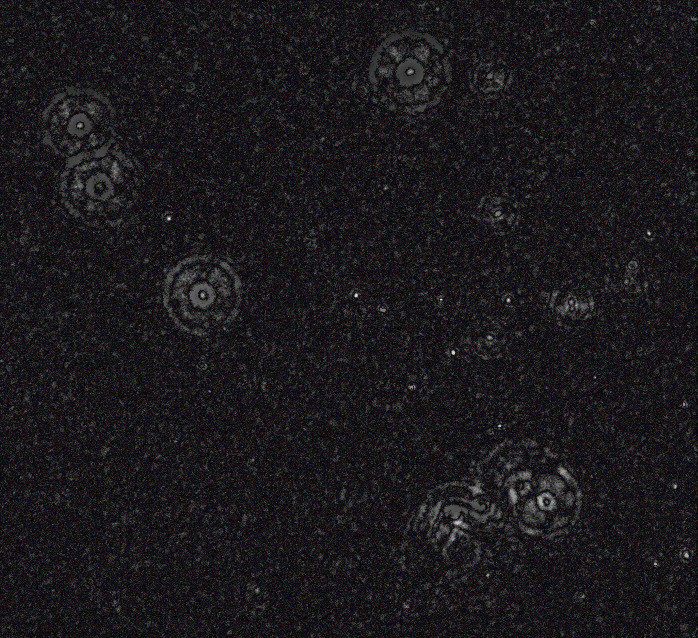
We could then copy that image, blur it and subtract to remove the low-frequency background features. Firstly with a small blur:
magick 1.jpg \( clone -blur 0x10 \) -compose difference -composite -auto-level 2.jpg
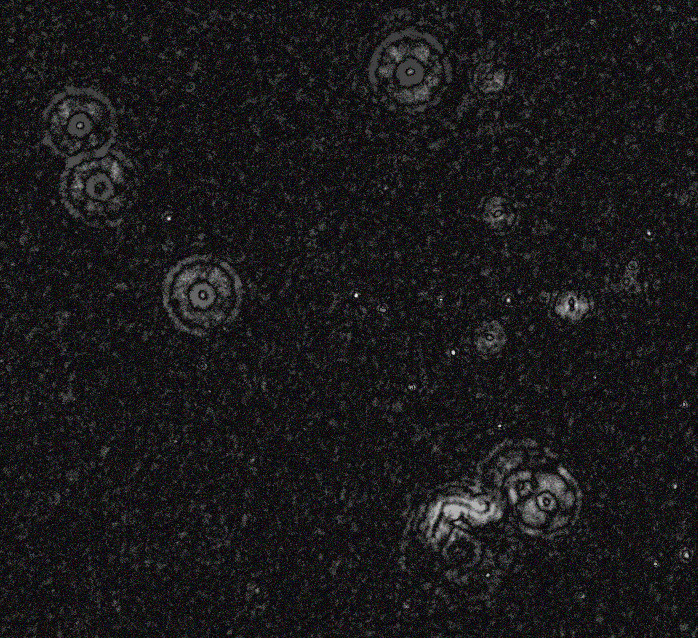
Then with a larger blur:
magick 1.jpg \( clone -blur 0x80 \) -compose difference -composite -auto-level 3.jpg
Or, we could go back to our original contrast-stretched image 1.jpg and try CLAHE on that:
magick 1.jpg -clahe 20x20% 128 20 result.jpg
Any image-processing wizards welcome to copy, steal, disagree, improve, extend, edit any concepts here...
CodePudding user response:
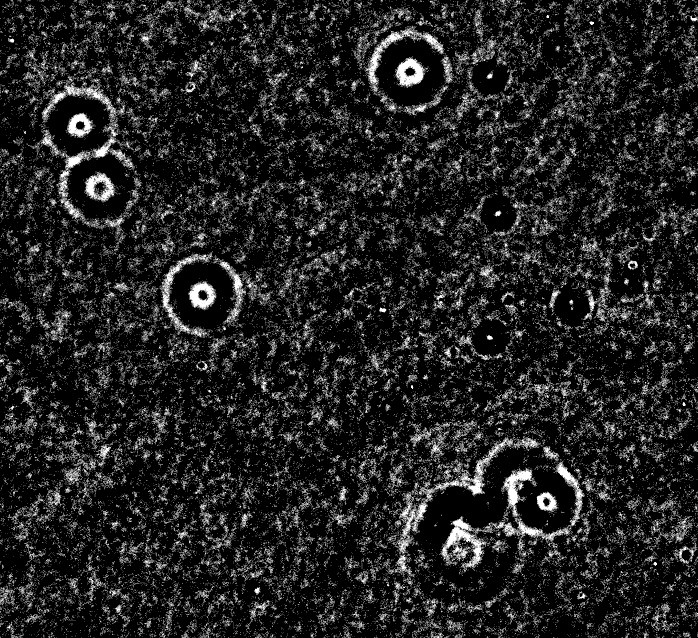
Here is another approach using division normalization followed by some dynamic range stretching in Python/OpenCV.
- Read the input
- Convert to grayscale
- Blur with relatively large sigma
- Divide the blurred image by the input
- Stretch the dynamic range
- Save the result
Input:
import cv2
import numpy as np
import skimage.exposure
# load image
img = cv2.imread("monocytes.png")
# convert to grayscale
gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
# blur
blur = cv2.GaussianBlur(gray, (0,0), sigmaX=39, sigmaY=39)
# divide
divide = cv2.divide(blur, gray, scale=255)
divide = 255 - divide
# stretch
maxval = np.amax(divide)/4
stretch = skimage.exposure.rescale_intensity(divide, in_range=(0,maxval), out_range=(0,255)).astype(np.uint8)
# write result to disk
cv2.imwrite("monocytes_division.png", divide)
cv2.imwrite("monocytes_stretch.jpg", stretch)
# display it
cv2.imshow("gray", gray)
cv2.imshow("divide", divide)
cv2.imshow("stretch", stretch)
cv2.waitKey(0)
cv2.destroyAllWindows()
Stretched Result: