I have a timeseries data which contain some peaks and valleys which significantly differ from the threshold level (an example vector is provided below).
The peak/valley height/width may vary as well as the noise level.
I am interested in finding & reporting both peaks and valleys.
Currently I am using a function based on this thread:
CodePudding user response:
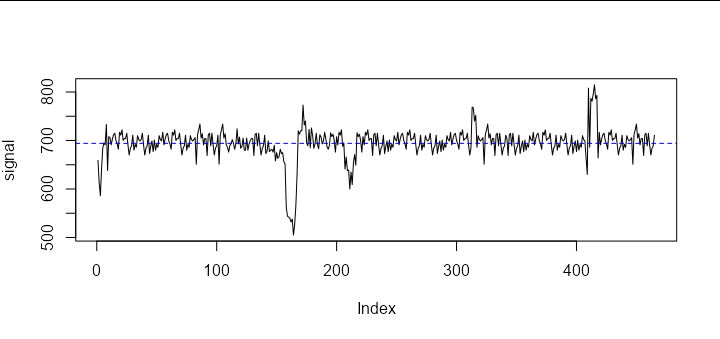
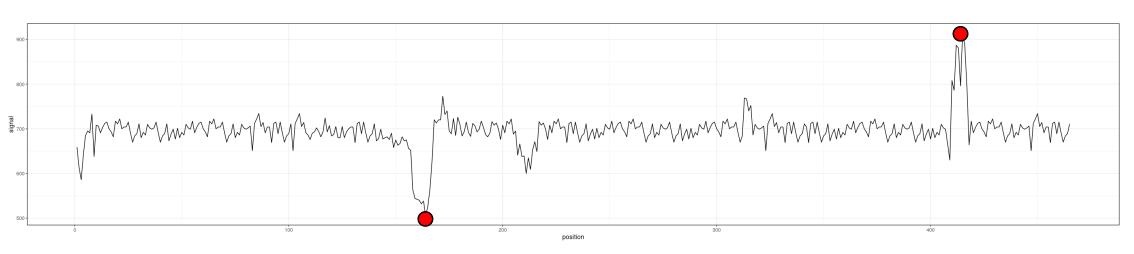
Let's start by plotting your data, along with its mean:
plot(signal, type = 'l')
abline(h = mean(signal), col = 'blue3', lty = 2)
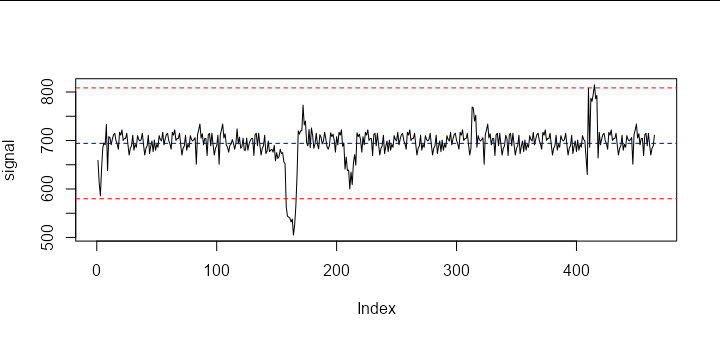
Since you want the upwards peak near the end of the series to be included but not the downwards spike at the very start, we need to find the number of standard deviations that would identify this peak but not the one at the start. This turns out to be about 3.5 standard deviations, as we can see if we plot the 3.5 standard deviation lines:
abline(h = mean(signal) c(3.5, -3.5) * sd(signal), lty = 2, col = 'red')
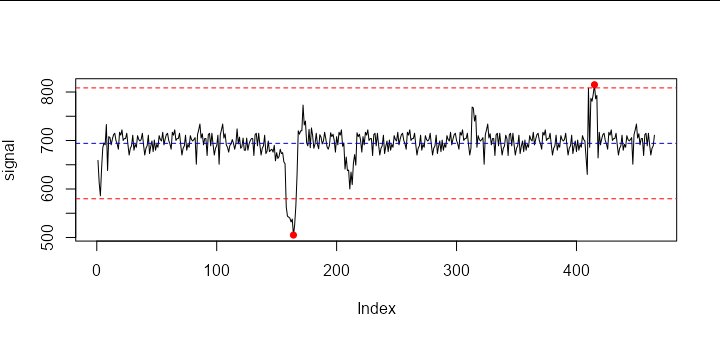
Now comes the tricky part. We use run length encoding to identify which contiguous parts of the sequence are outside of the 3.5 standard deviations, and find the point of largest absolute deviation for each:
exceed <- split(seq_along(big), data.table::rleid(big))[rle(big)$value]
peaks <- sapply(exceed, function(x) x[which.max(abs(signal[x] - mean(signal)))])
Now the vector peaks will contain only the maximum or minimum point within each spike. Note although you only have two spikes in your sample data, this will work for however many spikes you have.
To demonstrate, let us plot the points:
points(peaks, signal[peaks], col = 'red', pch = 16)
CodePudding user response:
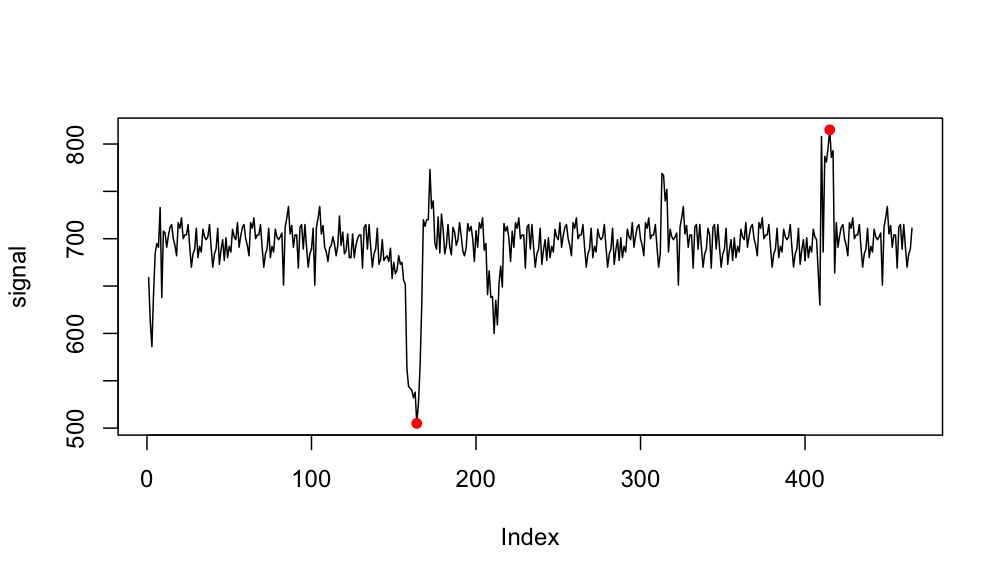
Maybe you want something like this:
max.index <- which.max(signal)
max <- max(signal)
min.index <- which.min(signal)
min <- min(signal)
plot(signal, type = "l")
points(max.index, max, col = "red", pch = 16)
points(min.index, min, col = "red", pch = 16)
Output: