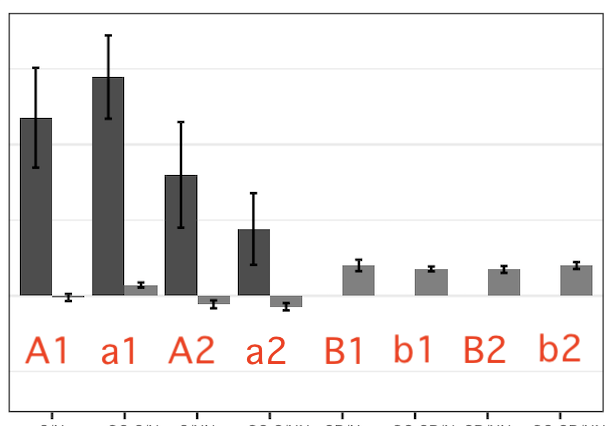
I have a dataset that contains multiple groups and exp. conditions. (here represents as group <- c("g1", "g2") and con <- c("A1", "A2", "B1", "B2")) I made a 3x3 plot matrix, and below is a part of one of the plots. a1 in the graph is just a derivative con of A1 and do not need to have a separate background than A1.
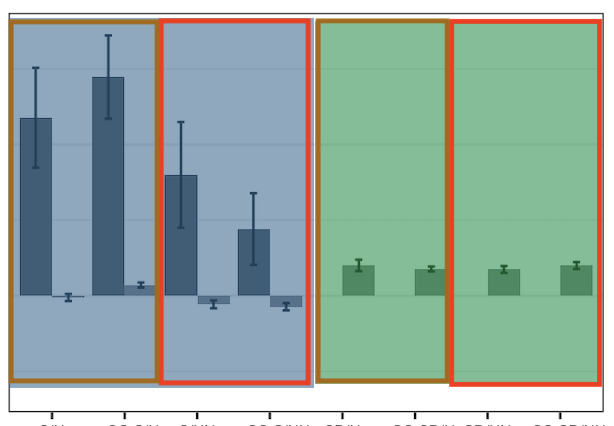
What I want is give them different colour background and frames so they are more friendly to readers. (e.g. the following output but fill the entire plating area [colour filled background distinguishing condition A, B, while coloured border differentiates condition 1, 2]) How could I archive this in R?
---- Update ----
As you commented, Here is a demo data that preserves the structure of the original data frame.
(sep = " ")# I use csv in real data,
# here is just for better presentation
group animal_nr region con gene1 gene2 gene3
g1 101 x A1 10 15 100
g1 102 x A1 12 15 110
g1 103 x A1 9 16 90
g1 104 x A1 11 14 105
g1 201 x a1 10 15 100
g1 202 x a1 12 16 110
g1 203 x a1 9 16 90
g1 204 x a1 11 14 105
g1 105 x A2 -8 5 50
g1 106 x A2 -9 5.5 52
g1 107 x A2 -8 4 53
g1 108 x A2 0 6 52.8
g1 205 x a2 -8 5 50
g1 206 x a2 -9 5.5 52
g1 207 x a2 -8 4 53
g1 208 x a2 0 6 52.8
g1 109 x B1 2 15 29
g1 110 x B1 3 15.5 35
g1 111 x B1 1 16 33
g1 112 x B1 1.1 14 30
g1 209 x b1 2 15 29
g1 210 x b1 3 15.5 35
g1 211 x b1 1 16 33
g1 212 x b1 1.1 14 30
g1 113 x B2 -10 2 10
g1 114 x B2 -8 3 11
g1 115 x B2 -9 2 11.1
g1 116 x B2 -11 4 12
g2 301 x A1 10 15 100
g2 302 x A1 12 15 110
g2 303 x A1 9 16 90
g2 304 x A1 11 14 105
g2 401 x a1 10 15 100
g2 402 x a1 12 16 110
g2 403 x a1 9 16 90
g2 404 x a1 11 14 105
g2 305 x A2 -8 5 50
g2 306 x A2 -9 5.5 52
g2 307 x A2 -8 4 53
g2 308 x A2 0 6 52.8
g2 405 x a2 -8 5 50
g2 406 x a2 -9 5.5 52
g2 407 x a2 -8 4 53
g2 408 x a2 0 6 52.8
Layout of real plot
grid(r = 3, n = 3)
gene/region x y z
gene1
gene2
gene3
CodePudding user response:
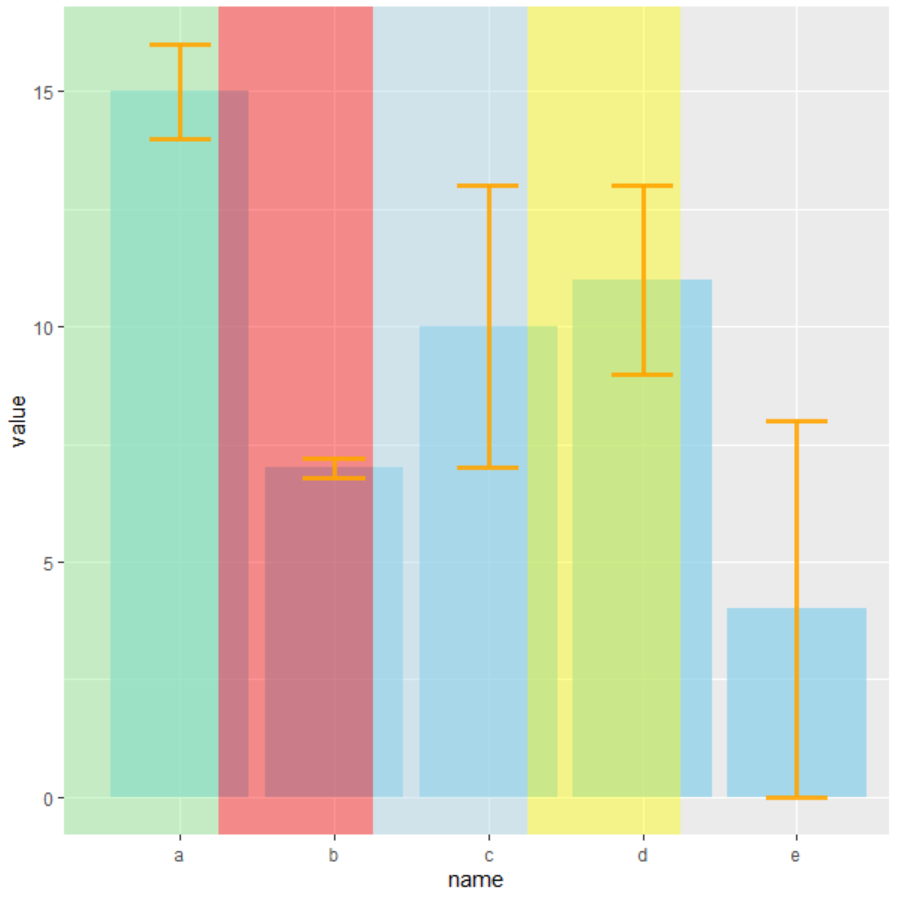
Here you have an example with dummy data and using geom_rec you can select that part of the background that you want to fill with some color, changing xmin and xmax.
library(tidyverse)
# create dummy data
data <- data.frame(
name=letters[1:5],
value=sample(seq(4,15),5),
sd=c(1,0.2,3,2,4)
)
# Plot
ggplot(data)
geom_bar( aes(x=name, y=value), stat="identity", fill="skyblue", alpha=0.7)
geom_rect(data=NULL,aes(xmin=0.25,xmax=1.25,ymin=-Inf,ymax=Inf),
fill="lightgreen", alpha=0.1)
geom_rect(data=NULL,aes(xmin=1.25,xmax=2.25,ymin=-Inf,ymax=Inf),
fill="red", alpha=0.1)
geom_rect(data=NULL,aes(xmin=2.25,xmax=3.25,ymin=-Inf,ymax=Inf),
fill="lightblue", alpha=0.1)
geom_rect(data=NULL,aes(xmin=3.25,xmax=4.25,ymin=-Inf,ymax=Inf),
fill="yellow", alpha=0.1)
geom_errorbar(aes(x=name, ymin=value-sd, ymax=value sd),
width=0.4, colour="orange", alpha=0.9, size=1.3)
CodePudding user response:
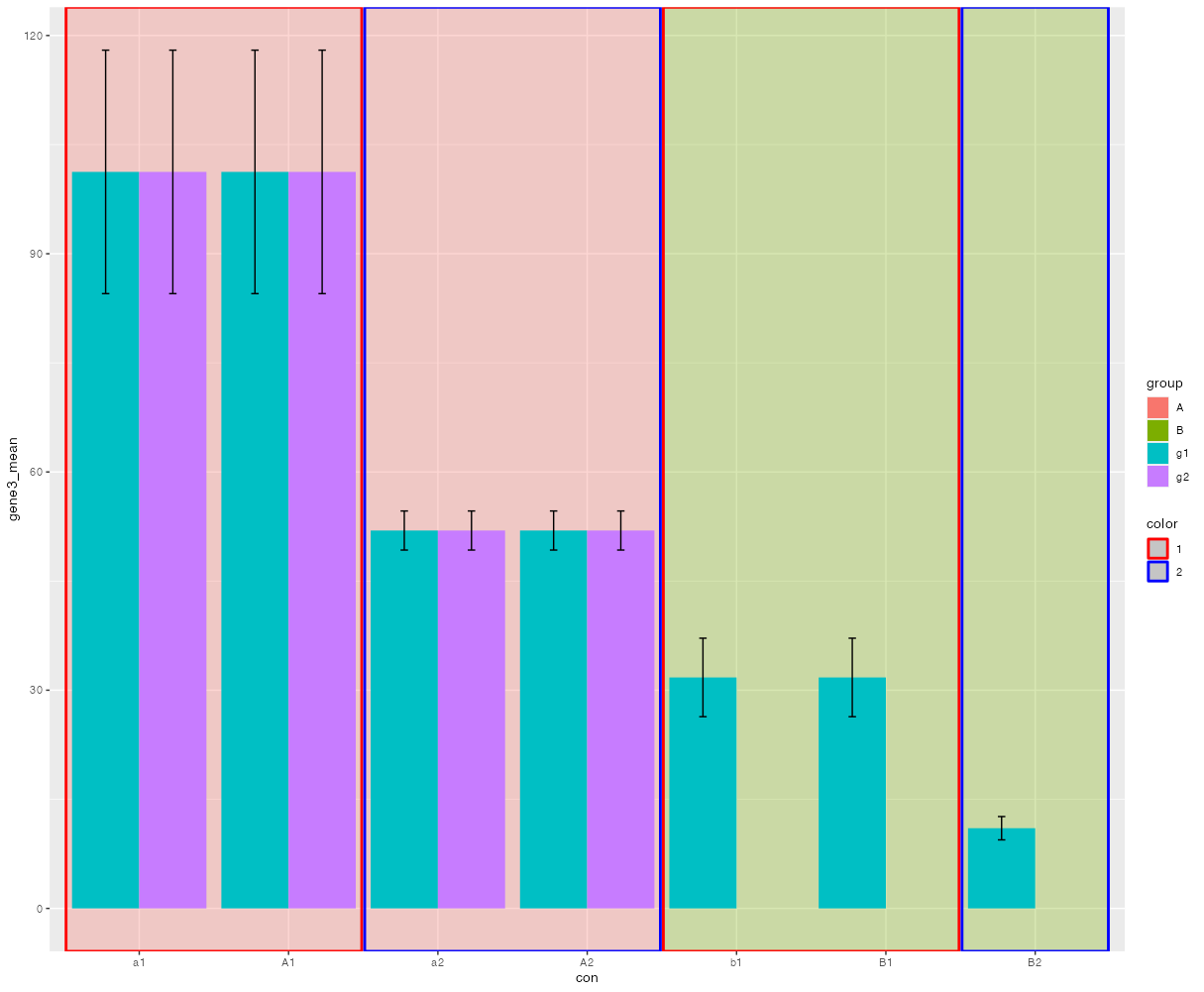
In general you could add some background rectangles using geom_rect. which however requires some data wrangling to get the xmin and xmax values. Basically this involves setting up a dataframe and converting your categories to numeric positions. Also at this stage we also add some columns which could be mapped on fill and color to achieve your desired result. Note that I slightly reduced the rect size to width_rect = .98 to prevent the outlines from overlapping.
library(dplyr)
library(ggplot2)
# Prep data for column and errorbars
df_sum <- df |>
group_by(group, con) |>
summarise(across(gene3, .fns = list(mean = mean, sd = sd)))
# Set up dataframe for background rectangles
width_rect <- .98
rects <- data.frame(
con = factor(unique(df$con))
)
rects <- rects |>
mutate(con_num = as.numeric(con),
con_group = toupper(con)) |>
group_by(con_group) |>
summarise(xmin = min(con_num) - width_rect / 2,
xmax = max(con_num) width_rect / 2) |>
mutate(fill = substr(con_group, 1, 1),
color = substr(con_group, 2, 2))
pd <- position_dodge(width = .9, preserve = "single")
ggplot(df_sum, aes(con, gene3_mean, fill = group))
scale_x_discrete()
geom_rect(aes(xmin = xmin, xmax = xmax, fill = fill, color = color, ymin = -Inf, ymax = Inf),
data = rects, inherit.aes = FALSE, size = 1, alpha = .3)
geom_col(position = pd)
geom_errorbar(aes(ymin = gene3_mean - 1.96 * gene3_sd,
ymax = gene3_mean 1.96 * gene3_sd),
width = .1, position = pd)
scale_color_manual(values = c("red", "blue"))