How can I regress on the previous time step in R here? I want to regress deaths at time t based on deaths, guns, and shootings at time t-1.
City Year Month Deaths Guns Shootings
Miami 2010 1 69 73800 701
Miami 2010 2 99 85050 738
Miami 2010 3 122 92650 784
...
Miami 2013 5 204 99280 800
Miami 2013 6 234 110023 825
Houston 2011 1 98 92100 789
Houston 2011 2 146 103900 799
Houston 2011 3 162 136100 772
Also in R, how do I use ggplot to show the month AND the year on the x-axis? I had a numeric date column in my dataset as well but I couldn't get it to plot by month and year.
CodePudding user response:
You can use dplyr::lag to lag the data and then fit the linear model. And, since a dplyr function is being used, I will do it in a pipe.
x <- 'City Year Month Deaths Guns Shootings
Miami 2010 1 69 73800 701
Miami 2010 2 99 85050 738
Miami 2010 3 122 92650 784
Miami 2013 5 204 99280 800
Miami 2013 6 234 110023 825
Houston 2011 1 98 92100 789
Houston 2011 2 146 103900 799
Houston 2011 3 162 136100 772'
df1 <- read.table(textConnection(x), header = TRUE)
suppressPackageStartupMessages(library(dplyr))
fit <- df1 %>%
mutate(Guns = lag(Guns), Shootings = lag(Shootings)) %>%
lm(Deaths ~ lag(Deaths) Guns Shootings, .)
summary(fit)
#>
#> Call:
#> lm(formula = Deaths ~ lag(Deaths) Guns Shootings, data = .)
#>
#> Residuals:
#> 2 3 4 5 6 7 8
#> -17.2273 0.8505 29.1597 53.4768 -63.1397 -38.9155 35.7956
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -9.668e 02 9.909e 02 -0.976 0.401
#> lag(Deaths) 2.567e-01 8.452e-01 0.304 0.781
#> Guns -8.023e-03 8.976e-03 -0.894 0.437
#> Shootings 2.364e 00 2.152e 00 1.099 0.352
#>
#> Residual standard error: 59.97 on 3 degrees of freedom
#> (1 observation deleted due to missingness)
#> Multiple R-squared: 0.3335, Adjusted R-squared: -0.333
#> F-statistic: 0.5004 on 3 and 3 DF, p-value: 0.708
Created on 2022-07-02 by the reprex package (v2.0.1)
CodePudding user response:
You could convert the year and month to a date object using clock's date_build to ggplot the Deaths or fitted values etc. against the date.
And if you wanted separate lm models per City you could nest by City and iterate through the models using purrr's map.
library(tidyverse)
library(clock)
library(broom)
# With some further made-up data
df <- tribble(
~City, ~Year, ~Month, ~Deaths, ~Guns, ~Shootings,
"Miami", 2010L, 1L, 69, 73800, 701,
"Miami", 2010L, 2L, 99, 85050, 738,
"Miami", 2010L, 3L, 122, 92650, 784,
"Miami", 2010L, 4L, 204, 99280, 800,
"Miami", 2010L, 5L, 234, 110023, 825,
"Miami", 2010L, 6L, 244, 110500, 830,
"Houston", 2011L, 1L, 98, 92100, 789,
"Houston", 2011L, 2L, 146, 103900, 799,
"Houston", 2011L, 3L, 162, 136100, 772,
"Houston", 2011L, 4L, 182, 146100, 782,
"Houston", 2011L, 5L, 192, 156100, 792
)
df2 <- df |>
group_by(City) |>
mutate(date = date_build(Year, Month),
lead_deaths = lead(Deaths)) |>
drop_na() |>
nest() |>
mutate(model = map(data, ~lm(lead_deaths ~ Deaths Guns Shootings, data = .)),
augmented = map(model, augment),
fitted = map(augmented, ".fitted")) |>
unnest(c(data, fitted)) |>
ungroup()
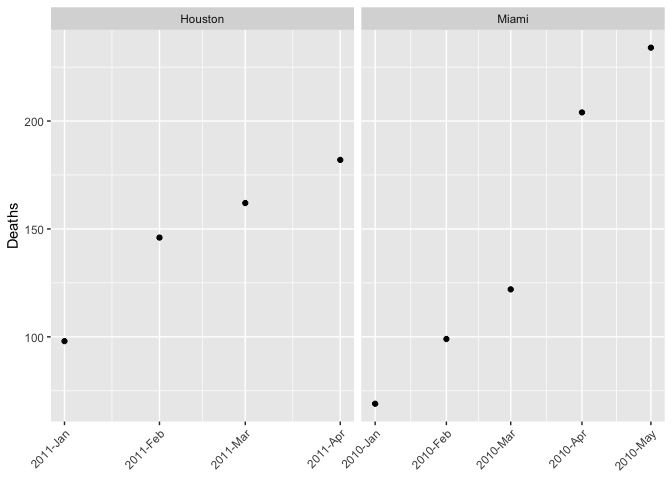
df2 |>
ggplot(aes(date, Deaths))
geom_point()
scale_x_date(date_labels = "%Y-%b")
facet_wrap(~ City, scales = "free_x")
labs(x = NULL)
theme(axis.text.x = element_text(angle = 45, hjust = 1))
Created on 2022-07-02 by the reprex package (v2.0.1)
