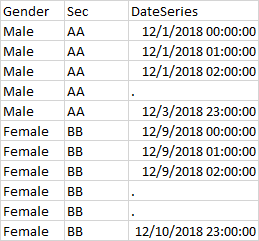
Hi I have a table df_in with 4 columns as shown below.I am getting error while trying to create dateseries on hourly basis with group by few columns.
Gender = c("Male","Female")
Sec = c("AA","BB")
min_date = c("12/1/2018","12/9/2018")
max_date = c("12/3/2018","12/10/2018")
df_in = data.frame(Gender,Sec,min_date,max_date)
for 1st row number of rows will be 72 (3 days * 24 hours) and for 2nd row 48 rows (2 days * 24 hours)
df_out will look like
I have tried with below code
df_out = df_in %>% group_by(Gender, Sec) %>% mutate (DateSeries = seq(as.POSIXct(min_date), as.POSIXct(max_date), by="hour"))
Thanks in advance.
CodePudding user response:
Does this produce what you need?
df_in %>%
group_by(Gender, Sec) %>%
expand(
nesting(Gender,Sec),
DateSeries = seq(as.POSIXct(min_date), as.POSIXct(max_date), by="hour")
)
# A tibble: 2,162 x 3
# Groups: Gender, Sec [2]
Gender Sec DateSeries
<chr> <chr> <dttm>
1 Female BB 0012-09-20 00:00:00
2 Female BB 0012-09-20 01:00:00
3 Female BB 0012-09-20 02:00:00
4 Female BB 0012-09-20 03:00:00
5 Female BB 0012-09-20 04:00:00
6 Female BB 0012-09-20 05:00:00
7 Female BB 0012-09-20 06:00:00
8 Female BB 0012-09-20 07:00:00
9 Female BB 0012-09-20 08:00:00
10 Female BB 0012-09-20 09:00:00
# ... with 2,152 more rows
Your first effort with mutate won't work because you're trying to add a 721 element vector to a data frame with only one row. expand is the function you need: it creates and "all possible combinations" version of its inputs.