I am trying to get annotations from a Tukey's test on to a facet'ed ggplot. I've tried multiple other answers (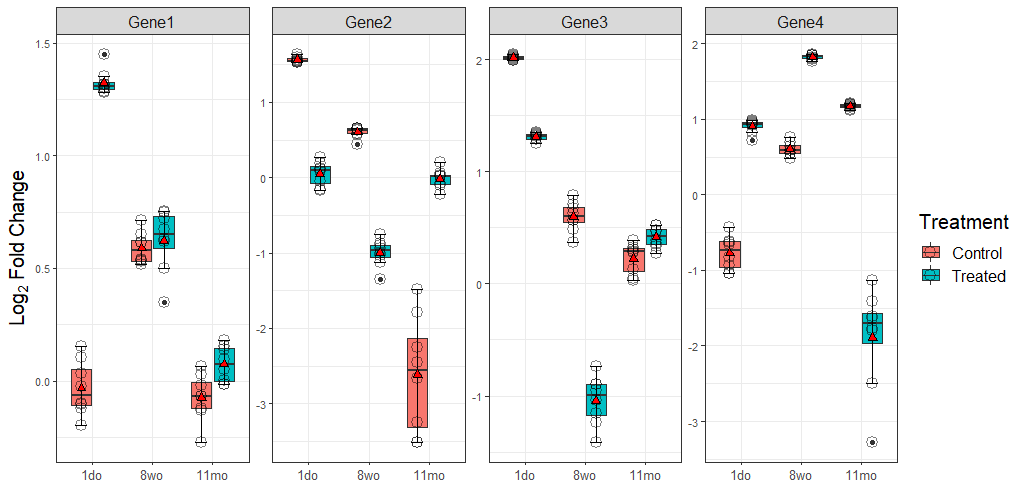
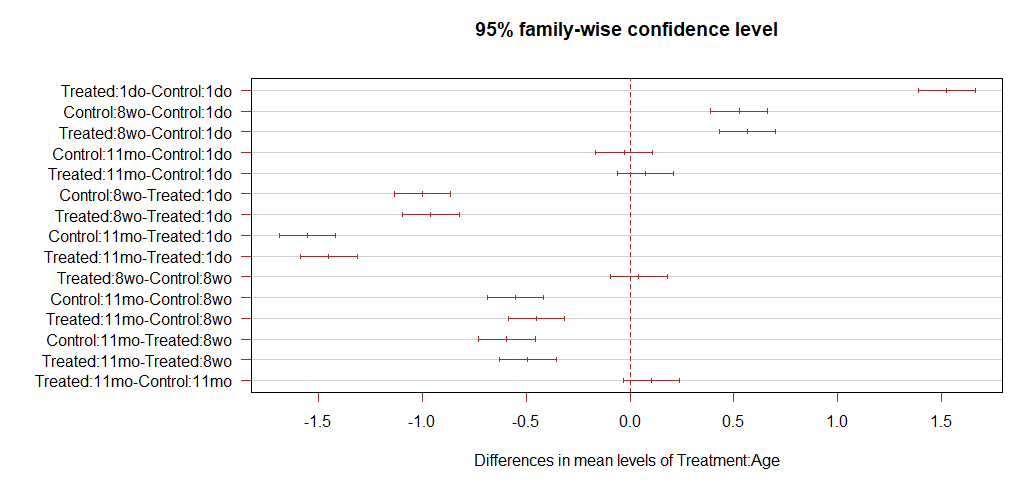
I can get the confidence interval plots one gene at a time using:
test_stats_Tukey <- Raw_data %>%
nest_by(Gene) %>%
mutate(Model = list(glm(FC ~ Treatment * Age, data = data)))
par(mar=c(5,13,4,1) .1)
plot(TukeyHSD(x= aov(test_stats_Tukey[[3]][[1]]), conf.level=0.95) , las=1 , col="brown")
Which gives:
But I'd like to extract and plot annotations in facets automatically.
Any help would be great. Thank you!
CodePudding user response:
It's not pretty, but it gets the job done!
Final_ann <- data.frame()
for (ann_facet in as.character(unique(Raw_data$Gene))) {
temp_ann_data <- Raw_data[which(Raw_data$Gene == ann_facet),]
temp_ann_model <- glm(FC ~ Treatment * Age,
data = temp_ann_data)
temp_ann_pairs <- emmeans(temp_ann_model,
pairwise ~ Treatment*Age,
adjust = "tukey")
temp_ann_model_cld <- cld(temp_ann_pairs$emmeans,
alpha = .05,
Letters = letters)
temp_ann_letters <- arrange(temp_ann_model_cld, Age, Treatment)
colnames(temp_ann_letters)[8] <- "Annotation"
temp_ann_letters$Annotation <- gsub("[[:space:]]", "",as.character(temp_ann_letters$Annotation))
temp_ann_letters$Gene <- ann_facet
temp_ann_letters$FC <-1.25*max(temp_ann_data$FC)
Final_ann <- rbind(Final_ann, temp_ann_letters[,c(1,2,8,9,10)])
}
Test_plot
geom_text(data = Final_ann,
aes(x = Age, y = log2(FC), group = Treatment, label=Annotation),
size=3, position = position_dodge(width = 0.75))
facet_wrap(Gene ~ ., ncol = 4, scales = "free")
CodePudding user response: