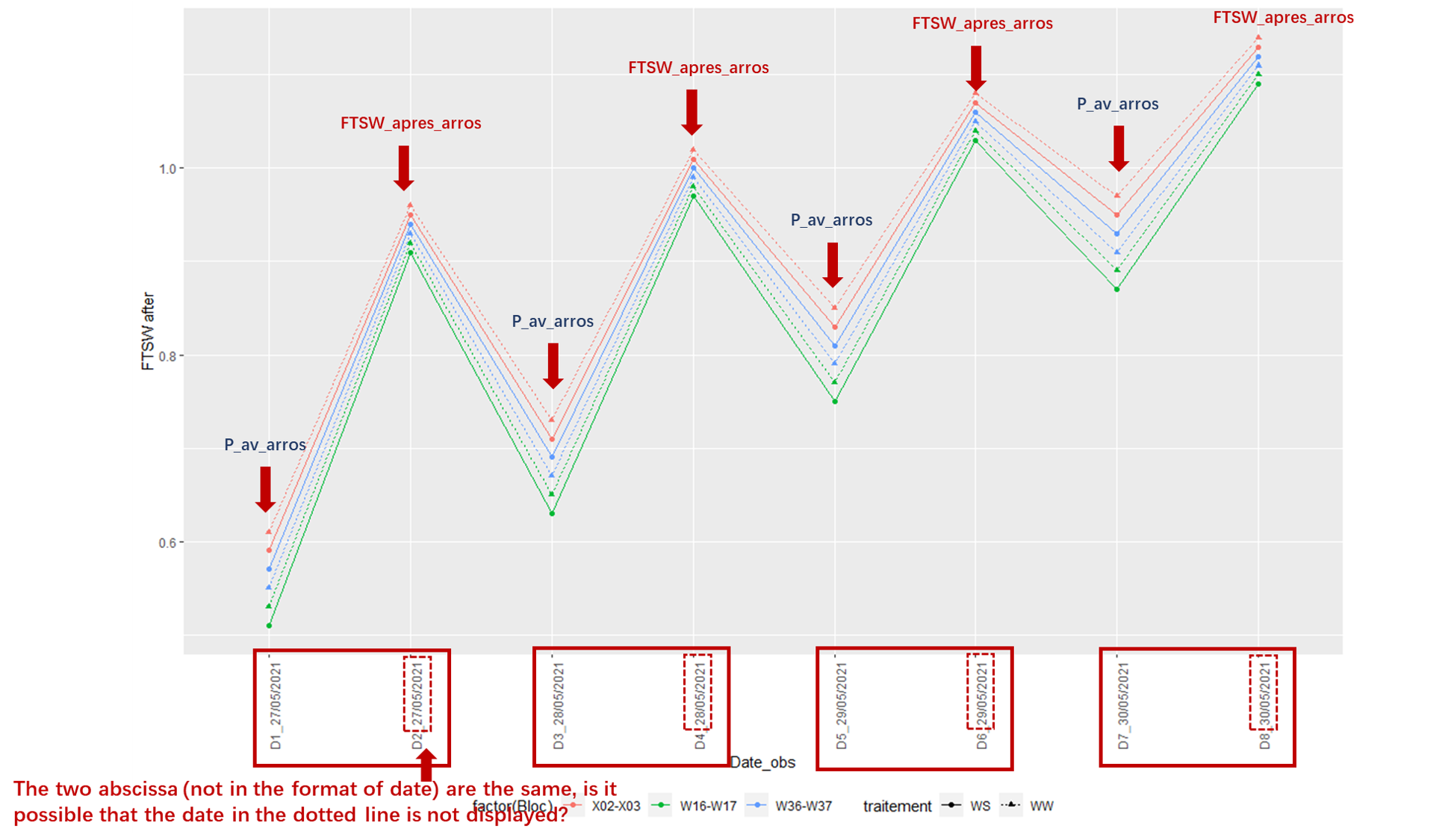
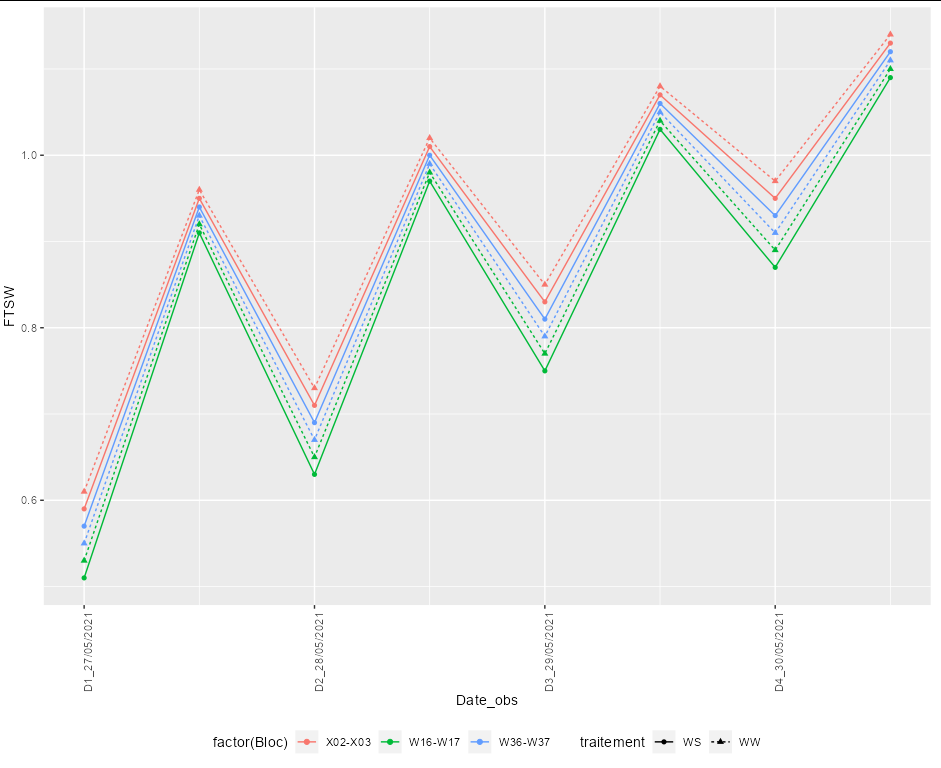
I want to plot two variables of the same abscissa in a same line. If it is the same abscissa, the two variables will be displayed on the same abscissa. But I want to keep them separate without affecting the coherence of the abscissa.
Here is the original data:
df<-structure(list(Bloc = c(7, 7, 8, 8, 5, 5, 7, 7, 8, 8, 5, 5, 7,
7, 8, 8, 5, 5, 7, 7, 8, 8, 5, 5), Pos_heliaphen = c("W16", "W17",
"W36", "W37", "X02", "X03", "W16", "W17", "W36", "W37", "X02",
"X03", "W16", "W17", "W36", "W37", "X02", "X03", "W16", "W17",
"W36", "W37", "X02", "X03"), traitement = c("WS", "WW", "WW",
"WS", "WS", "WW", "WS", "WW", "WW", "WS", "WS", "WW", "WS", "WW",
"WW", "WS", "WS", "WW", "WS", "WW", "WW", "WS", "WS", "WW"),
Variete = c("Blancas", "Blancas", "Blancas", "Blancas", "Blancas",
"Blancas", "Blancas", "Blancas", "Blancas", "Blancas", "Blancas",
"Blancas", "Blancas", "Blancas", "Blancas", "Blancas", "Blancas",
"Blancas", "Blancas", "Blancas", "Blancas", "Blancas", "Blancas",
"Blancas"), Date_obs = c("D1_27/05/2021", "D1_27/05/2021",
"D1_27/05/2021", "D1_27/05/2021", "D1_27/05/2021", "D1_27/05/2021",
"D2_28/05/2021", "D2_28/05/2021", "D2_28/05/2021", "D2_28/05/2021",
"D2_28/05/2021", "D2_28/05/2021", "D3_29/05/2021", "D3_29/05/2021",
"D3_29/05/2021", "D3_29/05/2021", "D3_29/05/2021", "D3_29/05/2021",
"D4_30/05/2021", "D4_30/05/2021", "D4_30/05/2021", "D4_30/05/2021",
"D4_30/05/2021", "D4_30/05/2021"), P_av_arros = c(0.51, 0.53,
0.55, 0.57, 0.59, 0.61, 0.63, 0.65, 0.67, 0.69, 0.71, 0.73,
0.75, 0.77, 0.79, 0.81, 0.83, 0.85, 0.87, 0.89, 0.91, 0.93,
0.95, 0.97), FTSW_apres_arros = c(0.91, 0.92, 0.93, 0.94,
0.95, 0.96, 0.97, 0.98, 0.99, 1, 1.01, 1.02, 1.03, 1.04,
1.05, 1.06, 1.07, 1.08, 1.09, 1.1, 1.11, 1.12, 1.13, 1.14
)), class = c("tbl_df", "tbl", "data.frame"), row.names = c(NA,
-24L))
I gained this code from previous questions. But this is when the abscissa is in the format of the date. Now I'm wondering what should I do if the abscissa is not a date, just a non-consecutive value?
library(ggplot2)
library(dplyr)
labels <- df %>%
select(Bloc, Pos_heliaphen) %>%
distinct(Bloc, Pos_heliaphen) %>%
group_by(Bloc) %>%
summarise(Pos_heliaphen = paste(Pos_heliaphen, collapse = "-")) %>%
tibble::deframe()
df %>%
mutate(Date_obs = as.POSIXct(lubridate::dmy(Date_obs))) %>%
tidyr::pivot_longer(6:7) %>%
mutate(Date_obs = if_else(name == "FTSW_apres_arros",
Date_obs 43200, Date_obs)) %>%
ggplot(aes(Date_obs, value, colour = factor(Bloc), shape = traitement,
linetype = traitement, group = interaction(Bloc, traitement)))
geom_point()
geom_line()
scale_color_discrete(labels = labels, guide = guide_legend(order = 1))
scale_x_datetime(date_labels = "%d/%m/%Y", date_breaks = "day")
labs(y = expression(paste("FTSW")))
theme(legend.position = "bottom",
axis.text.x = element_text(angle = 90, hjust = 1))
Thank you in advance!
CodePudding user response:
In this particular case, it is going to be easiest to stick to dealing with "proper" dates and just recreating the labels for the x axis.
df %>%
tidyr::separate(Date_obs, c("Obs", "Date_obs"), sep = "_") %>%
mutate(Date_obs = as.POSIXct(lubridate::dmy(Date_obs))) %>%
tidyr::pivot_longer(7:8) %>%
mutate(Date_obs = if_else(name == "FTSW_apres_arros",
Date_obs 43200, Date_obs)) %>%
ggplot(aes(Date_obs, value, colour = factor(Bloc), shape = traitement,
linetype = traitement, group = interaction(Bloc, traitement)))
geom_point()
geom_line()
scale_color_discrete(labels = labels, guide = guide_legend(order = 1))
scale_x_datetime(labels = ~ paste0("D", seq_along(.x) - 1, "_",
strftime(.x, "%d/%m/%Y")),
date_breaks = "day")
labs(y = expression(paste("FTSW")))
theme(legend.position = "bottom",
axis.text.x = element_text(angle = 90, hjust = 1))