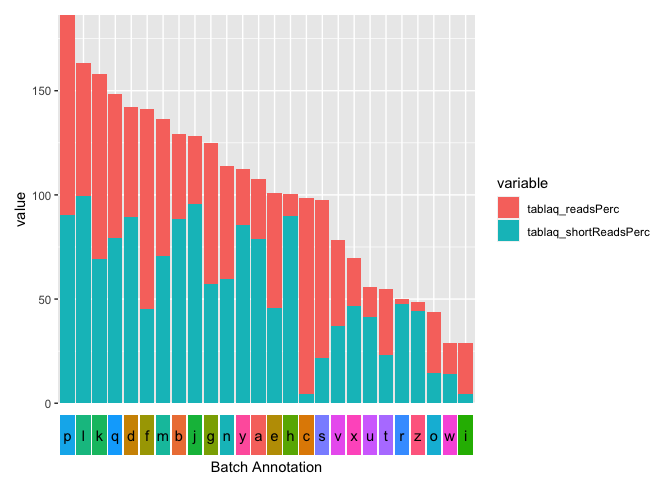
I'm building a barplot with RNA reads % in ggplot, I did this:
ggplot(tipos_exo,aes(x = reorder(sample, -value),y = value,fill = variable))
geom_bar( stat = "identity")
I need to replace the x axis labels with colored bars, each sample belongs to a specific batch and I looking for this effect:
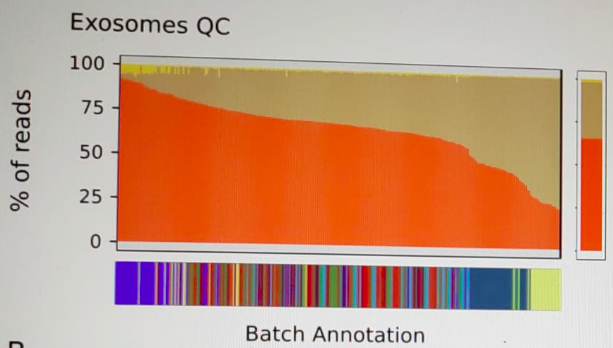
Any thoughts?
CodePudding user response:
One option to achieve your desired result would be to create your axis colorbar as a second plot and glue it to the main plot via the patchwork package.
For the colorbar I use geom_tile and remove all non-data ink using theme_void. As a first step I reorder your sample column by value and get rid of the duplicated sample categories using dplyr::distinct.
Using some fake random example data:
set.seed(123)
tipos_exo <- data.frame(
sample = rep(letters, each = 2),
variable = c("tablaq_readsPerc", "tablaq_shortReadsPerc"),
value = runif(52, 0, 100),
batch = rep(LETTERS, each = 2)
)
library(ggplot2)
library(patchwork)
library(dplyr, warn = FALSE)
p1 <- ggplot(tipos_exo,aes(x = reorder(sample, -value),y = value,fill = variable))
geom_bar( stat = "identity")
scale_y_continuous(expand = c(0, 0))
labs(x = NULL)
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.length.x = unit(0, "pt"))
tipos_exo1 <- tipos_exo |>
mutate(sample = reorder(sample, -value)) |>
distinct(sample, batch)
p_axis <- ggplot(tipos_exo1, aes(x = sample, y = factor(1), fill = batch))
geom_tile(width = .9)
geom_text(aes(label = sample))
theme_void()
theme(axis.title.x = element_text())
labs(x = "Batch Annotation")
guides(fill = "none")
p1 / p_axis plot_layout(heights = c(8, 1))