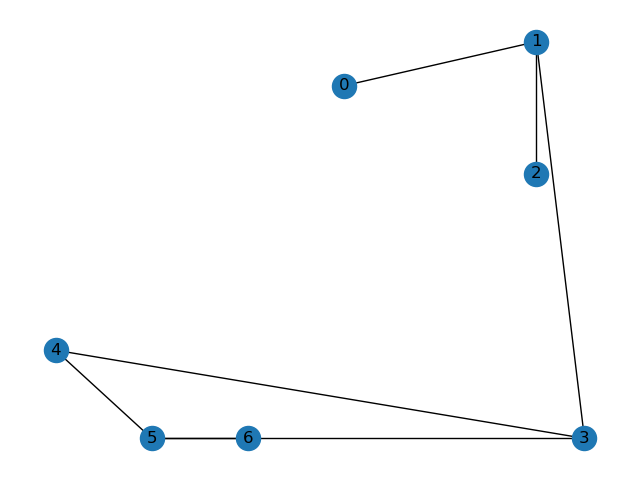
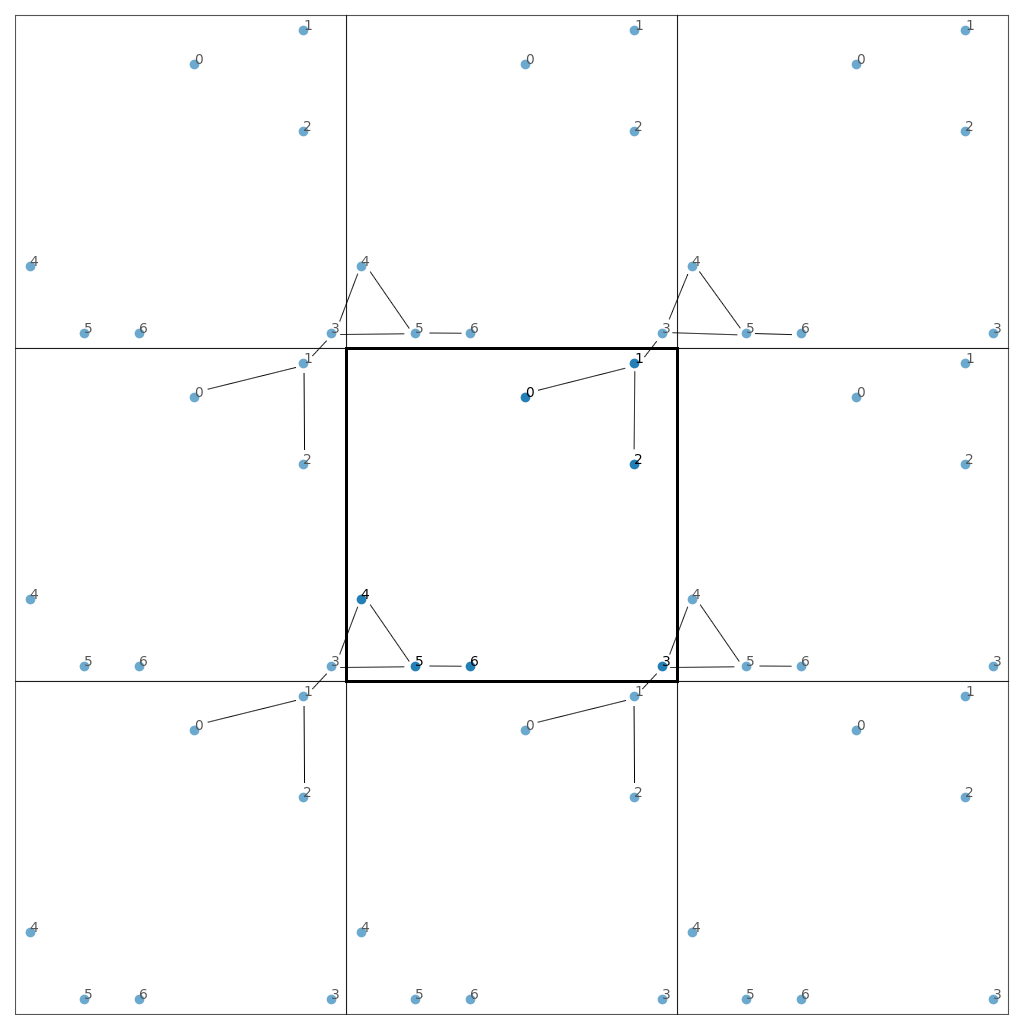
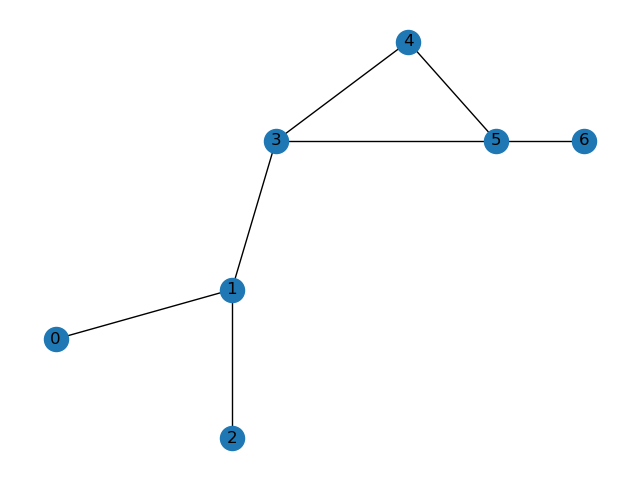
I have a "molecule" (which is really a graph consisting of 7 nodes) that is fragmented by periodic boundary conditions:
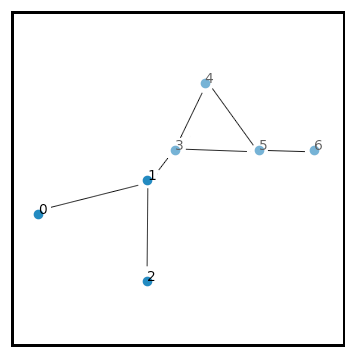
Essentially, the cell at the center is used to tile the entire space, and even though nodes 1 and 3 share an edge, they appear on the opposite sides of the cell at the center. I would like to unfragment this graph, to get:
Given
bonds = [(0,1),(1,2),(1,3),(3,4),(3,5),(4,5),(5,6)]
coords = np.array([[4,5],[6,5.5],[6,4],[6.5,1],[1,2],[2,1],[3,1]])
and cell dimensions
cell = [7,6]
I try the following:
import networkx as nx
import numpy as np
import matplotlib.pyplot as plt
G = nx.Graph()
G.add_edges_from(bonds)
node = 2
processed = []
moved = []
Q = [node]
new_xyz = []
new_xyz.append(coords[node,:])
indices = [node]
while len(Q)>0:
curr = Q[0]
ref_pos = coords[curr,:]
Q = Q[1:]
processed.append(curr)
for i in G.adj[curr]:
if i not in processed:
Q.append(i)
if i not in moved:
print("moving ",i, "(ref: ",curr,")")
pos = coords[i,:]
dist = pos - ref_pos
dist = nearest_image(dist,cell)
pos2 = dist ref_pos
new_xyz.append(pos2)
indices.append(i)
assert(curr in moved or curr==node)
moved.append(i)
new_xyz = np.array(new_xyz)
where
def nearest_image(d,cell):
for i in range(2):
hbox = cell[i] / 2
while d[i] > hbox:
d[i] -= cell[i]
while d[i] < -hbox:
d[i] = cell[i]
assert(d[i] < hbox and d[i] > -hbox)
return d
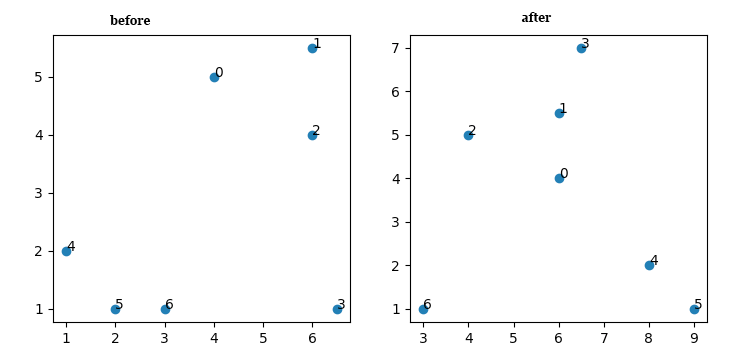
When I plot the results using:
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(5,3))
axes[0].scatter(coords[:,0],coords[:,1])
for i in range(7):
axes[0].annotate(str(i), (coords[i,0],coords[i,1]))
axes[1].scatter(new_xyz[:,0],new_xyz[:,1])
for i in indices:
axes[1].annotate(str(i), (new_xyz[i,0],new_xyz[i,1]))
plt.show()
I get:
What am I missing? I don't quite understand how 0 and 2 swap places either.