I have browsed multiple similar questions here but always get stuck by different problems, probably because I am not good at R. I need to create a forest plot with several variables made of various levels of factors that includes its uni- and multivariable HR estimates. The HR lines of each factor level should be on top of each other and grouped by the main variables.
I have tried the code below, but I would like to have the main variable names on the left of the plot, next or above each factor (Not on the right as from the code). Also, the factor levels need to be ordered specifically, for example: General childhood condition levels: Poor, Fair, Good, Very good, Excellent. Any help would be very much appreciated!
Here is a snippet of my dataset & ggplot script:
df <- data.frame(
label = c("Low", "Middle", "High", "Good", "Fair", "Poor", "Very good", "Excellent", "Low", "Middle", "High", "Good", "Fair", "Poor", "Very good", "Excellent"),
var_label = c("Education", "Education", "Education", "General health", "General health", "General health", "General health", "General health", "Education", "Education", "Education", "General health", "General health", "General health", "General health", "General health"),
reference = c("TRUE", "FALSE", "FALSE", "TRUE", "FALSE", "FALSE", "FALSE", "FALSE", "TRUE", "FALSE", "FALSE", "TRUE", "FALSE", "FALSE", "FALSE", "FALSE"),
estimate = c(1.00, 1.34, 1.67, 1.00, 0.82, 1.11, 0.85, 1.48, 1.00, 2.67, 3.49, 0.83, 1.00, 0.95, 1.34),
std.error = c(NA, 0.16, 0.20, NA, 0.16, 0.24, 0.18, 0.24, NA, 0.14, 0.18, NA, 0.16, 0.23, 0.18, 0.24),
conf.low = c(NA, 0.95, 1.10, NA, 0.60, 0.69, 0.60, 0.92, NA, 2.01, 2.45, NA, 0.61, 0.63, 0.66, 0.83),
conf.high = c(NA, 1.87, 2.52, NA, 1.11, 1.80, 1.21, 2.39, NA, 3.55, 4.98, NA, 1.13, 1.58, 1.35, 2.16),
p.value = c(NA, 0.09, 0.02, NA, 0.20, 0.67, 0.38, 0.11, NA, 0.00, 0.00, NA, 0.24, 1.00, 0.76, 0.23),
model = c("m", "m", "m", "m", "m", "m", "m", "m", "u", "u", "u", "u", "u", "u", "u", "u")
)
dotCOLS = c("#a6d8f0","#f9b282")
barCOLS = c("#008fd5","#de6b35")
forest <- ggplot(df, aes(x = estimate, xmax= conf.high, xmin = conf.low, y = label, color = model, fill = model))
geom_point(size = 3, shape = 18, position = position_dodge(width = 0.5))
geom_linerange(position = position_dodge(width = 0.5), size = 1)
geom_vline(xintercept = 1, size = 1)
geom_hline(yintercept = 0, size = 1)
facet_grid(var_label~., scales = "free_y", space = "free_y")
scale_alpha_identity()
scale_fill_manual(values=barCOLS)
scale_color_manual(values=dotCOLS)
scale_y_discrete(name="Variable groups")
scale_x_continuous(name="Hazard ratio", limits = c(0, 5))
geom_text(aes(label = var_label), x = Inf, y = Inf, hjust = 1, vjust = 4.5, check_overlap = TRUE, color = "darkgreen")
theme(panel.background = element_blank(),
panel.spacing = unit(0, "pt"),
axis.line.x.bottom = element_line(size = 1),
strip.background = element_blank(), strip.text = element_blank())
And here is a picture of the result:
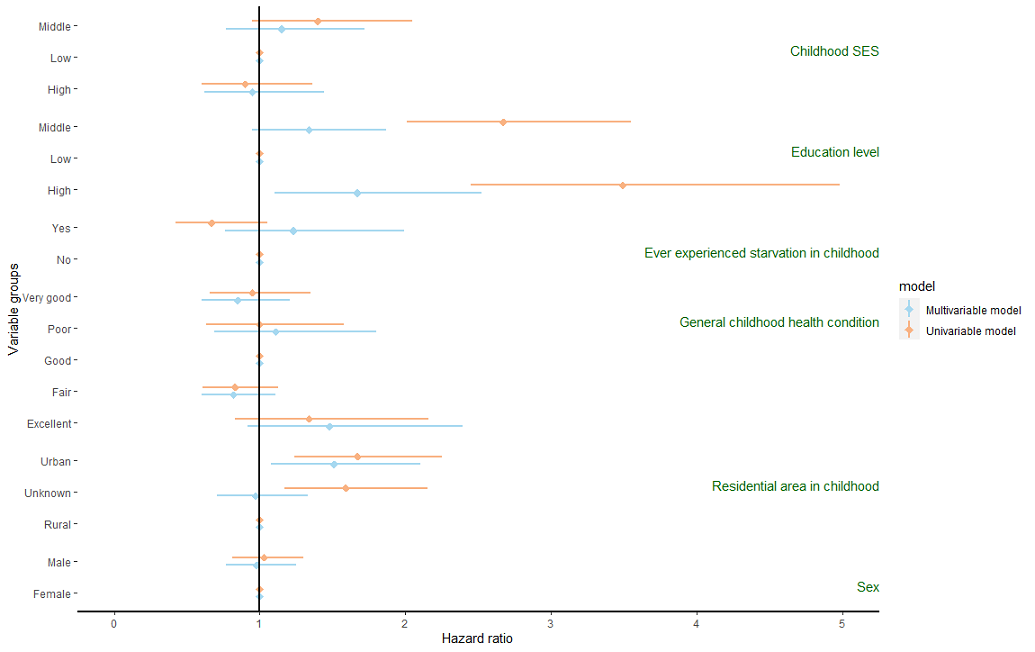
CodePudding user response:
As you want the labels on the left use x=-Inf in geom_text. Also, instead of shifting via vjust you could IMHO simply use vjust=1 to align at the top. Additionally use coord_cartesian(clip = "off") to prevent the labels from being clipped off and make some room for the labels by adding some margin to the y axis text. Finally, if you want your labels in a specific order than convert to a factor with the levels set in your desired order.
Note: I fixed your data which had only 15 values for the estimate variable while all other columns had a length of 16. Additionally I removed the geom_hline which IMHO is not necessary (at least for your example data).
dotCOLS <- c("#a6d8f0", "#f9b282")
barCOLS <- c("#008fd5", "#de6b35")
library(ggplot2)
df$label <- factor(df$label, levels = c("Low", "Middle", "High", "Good", "Fair", "Poor", "Very good",
"Excellent", "Very good"))
ggplot(df, aes(x = estimate, xmax = conf.high, xmin = conf.low, y = label, color = model, fill = model))
geom_point(size = 3, shape = 18, position = position_dodge(width = 0.5))
geom_linerange(position = position_dodge(width = 0.5), size = 1)
geom_vline(xintercept = 1, size = 1)
#geom_hline(yintercept = 0, size = 1)
facet_grid(var_label ~ ., scales = "free_y", space = "free_y")
scale_alpha_identity()
scale_fill_manual(values = barCOLS)
scale_color_manual(values = dotCOLS)
scale_y_discrete(name = "Variable groups")
scale_x_continuous(name = "Hazard ratio", limits = c(0, 5))
geom_text(aes(label = var_label), x = -Inf, y = Inf, hjust = 1, vjust = 1, check_overlap = TRUE, color = "darkgreen")
coord_cartesian(clip = "off")
theme(
panel.background = element_blank(),
panel.spacing = unit(0, "pt"),
axis.line.x.bottom = element_line(size = 1),
axis.text.y.left = element_text(margin = margin(l = 20, unit = "pt")),
strip.background = element_blank(),
strip.text = element_blank()
)
#> Warning: Removed 2 rows containing missing values (geom_segment).
#> Removed 2 rows containing missing values (geom_segment).
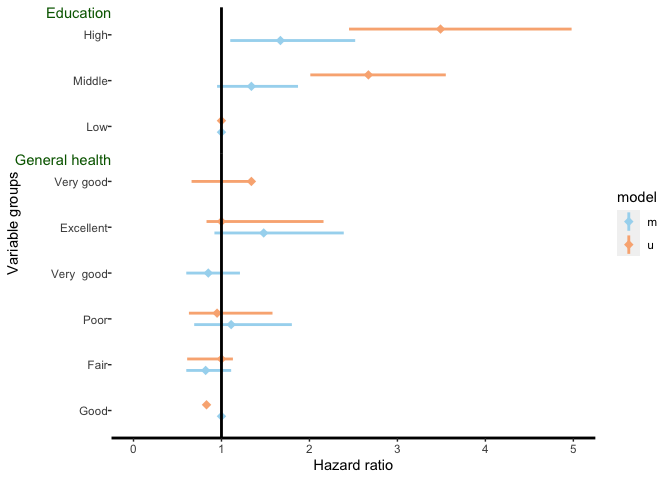
DATA
df <- data.frame(
label = c("Low", "Middle", "High", "Good", "Fair", "Poor", "Very good", "Excellent", "Low", "Middle", "High", "Good", "Fair", "Poor", "Very good", "Excellent"),
var_label = c("Education", "Education", "Education", "General health", "General health", "General health", "General health", "General health", "Education", "Education", "Education", "General health", "General health", "General health", "General health", "General health"),
reference = c("TRUE", "FALSE", "FALSE", "TRUE", "FALSE", "FALSE", "FALSE", "FALSE", "TRUE", "FALSE", "FALSE", "TRUE", "FALSE", "FALSE", "FALSE", "FALSE"),
estimate = c(1.00, 1.34, 1.67, 1.00, 0.82, 1.11, 0.85, 1.48, 1.00, 2.67, 3.49, 0.83, 1.00, 0.95, 1.34, 1),
std.error = c(NA, 0.16, 0.20, NA, 0.16, 0.24, 0.18, 0.24, NA, 0.14, 0.18, NA, 0.16, 0.23, 0.18, 0.24),
conf.low = c(NA, 0.95, 1.10, NA, 0.60, 0.69, 0.60, 0.92, NA, 2.01, 2.45, NA, 0.61, 0.63, 0.66, 0.83),
conf.high = c(NA, 1.87, 2.52, NA, 1.11, 1.80, 1.21, 2.39, NA, 3.55, 4.98, NA, 1.13, 1.58, 1.35, 2.16),
p.value = c(NA, 0.09, 0.02, NA, 0.20, 0.67, 0.38, 0.11, NA, 0.00, 0.00, NA, 0.24, 1.00, 0.76, 0.23),
model = c("m", "m", "m", "m", "m", "m", "m", "m", "u", "u", "u", "u", "u", "u", "u", "u")
)
