I have a problem where calling my current regular expression on very large strings seems to nuke a container I have the code running in. I know of one straightforward replacement for my code that is slightly slower, but I feel like there should be faster implementations. I would prefer to stay in base R.
An example of my code looks like:
# create a small example
library(microbenchmark)
set.seed(1986)
SET1 <- c("A", "B", "C", "D", "=", "E")
SET2 <- seq(1:100)
SET3 <- sample(x = 1:6,
size = 1000,
replace = TRUE)
CIGAR <- sapply(X = SET3,
FUN = function(x) {
z1 <- sample(x = SET1,
size = x,
replace = TRUE)
z2 <- sample(x = SET2,
size = x,
replace = TRUE)
z3 <- paste0(z1, z2, collapse = "")
},
simplify = TRUE)
# the original implementation:
# this appears to ask for exorbitant amounts of memory when the original string is very large
# or at least the docker container dies and that appears to be the cause
f1 <- function() {
CIGARType <- regmatches(x = CIGAR,
m = gregexpr(pattern = "[^0-9]",
text = CIGAR))
CIGARCount <- regmatches(x = CIGAR,
m = gregexpr(pattern = "[0-9] ",
text = CIGAR))
CIGARCount <- lapply(X = CIGARCount,
FUN = function(x) {
as.integer(x)
})
return(list(CIGARCount,
CIGARType))
}
# the current alternative:
f2 <- function() {
CIGARType <- CIGARCount <- vector(mode = "list",
length = length(CIGAR))
for (m1 in seq_along(CIGAR)) {
CIGARType[m1] <- regmatches(x = CIGAR[m1],
m = gregexpr(pattern = "[^0-9]",
text = CIGAR[m1]))
CIGARCount[m1] <- regmatches(x = CIGAR[m1],
m = gregexpr(pattern = "[0-9] ",
text = CIGAR[m1]))
CIGARCount[[m1]] <- as.integer(CIGARCount[[m1]])
}
return(list(CIGARCount,
CIGARType))
}
My alternative works and is slower, but the container doesn't explode:
> microbenchmark(f1() -> a, f2() -> b)
Unit: milliseconds
expr min lq mean median uq max neval
a <- f1() 13.84300 15.21332 20.20298 16.92589 19.15321 101.0486 100
b <- f2() 53.29438 58.50947 67.20698 62.89677 70.24507 158.4985 100
> all(mapply(function(x, y) all(x == y), x = a[[1]], y = b[[1]]))
[1] TRUE
> all(mapply(function(x, y) all(x == y), x = a[[2]], y = b[[2]]))
[1] TRUE
Are there alternative quick ways to parse these strings?
CodePudding user response:
Reduce the number of times you perform a regex, they are the most expensive component here. It is generally much faster to extract the non-number and number(s) in one step, then substring them. Try this:
f3 <- function() {
tmp <- regmatches(CIGAR, gregexpr("[^0-9][0-9] ", CIGAR))
types <- lapply(tmp, substr, 1, 1)
counts <- lapply(tmp, function(z) as.integer(substring(z, 2)))
list(counts, types)
}
out1 <- f1()
out3 <- f3()
identical(out1, out3)
# [1] TRUE
res <- bench::mark(f1(), f2(), f3(), min_iterations = 500)
res
# # A tibble: 3 x 13
# expression min median `itr/sec` mem_alloc `gc/sec` n_itr n_gc total_time result memory time gc
# <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl> <int> <dbl> <bch:tm> <list> <list> <list> <list>
# 1 f1() 11.13ms 15ms 63.6 15.9MB 52.4 274 226 4.31s <list [2]> <Rprofmem [4,587 x 3]> <bench_tm [500]> <tibble [500 x 3]>
# 2 f2() 43.03ms 50.4ms 18.8 15.82MB 39.3 162 338 8.6s <list [2]> <Rprofmem [4,002 x 3]> <bench_tm [500]> <tibble [500 x 3]>
# 3 f3() 8.15ms 10.1ms 99.0 7.96MB 29.2 386 114 3.9s <list [2]> <Rprofmem [2,008 x 3]> <bench_tm [500]> <tibble [500 x 3]>
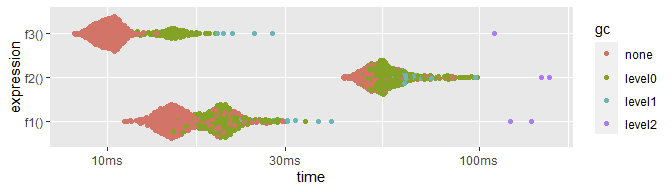
plot(res)
(Note the x-axis is log.)
The key column I look at is `itr/src`, which is saying that this operates 55% faster (in this run) than f1() and over 5x faster than f2(). Also note from mem_alloc that f3 is using just over half the memory allocation. (The actual performance ratios waffle between multiple runs, from 25-66% faster than f1, and 4.5-5.2x faster than f2. I'm showing just one. Every time I've done this benchmark, though, the relative performance is about the same, never does f3 come out anywhere worse than 26% faster than f1.)
(FYI, I hope your use of f1 with no arguments and scope-breaching for CIGAR is just contrived for the benchmark and/or this question. If you're writing functions that work on data, then pass the data as an argument to the function; never scope-breach like this if at all avoidable, it breaks reproducibility and can make troubleshooting frustratingly difficult.)