I have multiple distribution functions, lets say:
A <- function(x)dnorm(x, mean = 3, sd = 1)
B <- function(x)dnorm(x, mean = 6, sd = 1)
C <- function(x)dnorm(x, mean = 2, sd = 2)
I would like to create a new, proportional distribution function. For the three functions above, using the pdqr package, I can write this:
x <- seq(1, 10, by = 0.1)
P <- data.frame(x) %>%
mutate(A = A(x),
B = B(x),
C = C(x)) %>%
mutate(y = A * B * C) %>%
pdqr::new_d(type = "continuous")
This generates the proportional distribution P shown below, based on A, B and C.
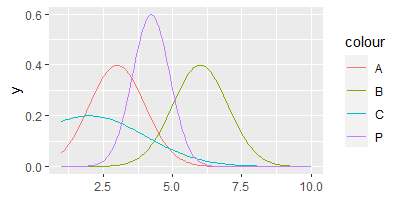
Now, since it is a drag to create new distribution functions based on a range of 2 to 10 other distributions, I would like to create a function, which takes a list of distributions to create a new proportional distribution function. Ideally, it would take this input: P <- foo(c(A, B, C))
However, I can't seem to get any further than:
foo <- function(list){
#set x values
x <- seq(1, 10, by = 0.1)
#create new function
data.frame(x) %>%
for(i in list){
mutate(i = i(x))%>%
mutate(y = i * i) %>%
new_d(type = "continuous")
}
}
Which obviously doesn't work. The question is twofold: how to pass the functions in the list to:
- create new vectors to calculate the propability for each
x - to create a
ywhich multiplies the values generated in step 1
CodePudding user response:
Some problems with using a for loop in a %>% pipe:
- The data is going nowhere, you would need to wrap your loop in
{-braces and use the.data placeholder. - Within the
forloop, you're calculating and then discarding the results. R doesn't magically know what that you intend to use it.
Frankly, your function may be clearer without dplyr/magrittr:
foo <- function(x, ...) {
y <- 1L # identity
for (fun in list(...)) y <- y * fun(x)
y
}
foo(x, A, B, C)
# [1] 1.413004e-08 2.884836e-08 5.758736e-08 1.123988e-07 2.144986e-07 4.002355e-07 7.301887e-07 1.302516e-06 2.271744e-06
# [10] 3.874040e-06 6.459473e-06 1.053073e-05 1.678604e-05 2.616173e-05 3.986695e-05 5.940021e-05 8.653490e-05 1.232602e-04
# [19] 1.716655e-04 2.337607e-04 3.112349e-04 4.051664e-04 5.157116e-04 6.418135e-04 7.809786e-04 9.291756e-04 1.080898e-03
# [28] 1.229420e-03 1.367238e-03 1.486676e-03 1.580581e-03 1.643031e-03 1.669948e-03 1.659543e-03 1.612511e-03 1.531952e-03
# [37] 1.423036e-03 1.292454e-03 1.147737e-03 9.965485e-04 8.460240e-04 7.022558e-04 5.699495e-04 4.522783e-04 3.509163e-04
# [46] 2.662133e-04 1.974623e-04 1.432080e-04 1.015496e-04 7.040736e-05 4.772941e-05 3.163607e-05 2.050252e-05 1.299153e-05
# [55] 8.048999e-06 4.875866e-06 2.887953e-06 1.672464e-06 9.470038e-07 5.242942e-07 2.838094e-07 1.502128e-07 7.773480e-08
# [64] 3.933258e-08 1.945887e-08 9.412636e-09 4.451776e-09 2.058655e-09 9.308129e-10 4.114997e-10 1.778710e-10 7.517422e-11
# [73] 3.106427e-11 1.255110e-11 4.958277e-12 1.915174e-12 7.232924e-13 2.670841e-13 9.642964e-14 3.404093e-14 1.174953e-14
# [82] 3.965229e-15 1.308411e-15 4.221324e-16 1.331623e-16 4.107168e-17 1.238603e-17 3.652160e-18 1.052922e-18 2.968046e-19
# [91] 8.180384e-20
which can be stored back into your frame. If you want to use it in a %>%-pipe, then
tibble(myx = x) %>%
mutate(y = foo(myx, A, B, C))
# # A tibble: 91 x 2
# myx y
# <dbl> <dbl>
# 1 1 0.0000000141
# 2 1.1 0.0000000288
# 3 1.2 0.0000000576
# 4 1.3 0.000000112
# 5 1.4 0.000000214
# 6 1.5 0.000000400
# 7 1.6 0.000000730
# 8 1.7 0.00000130
# 9 1.8 0.00000227
# 10 1.9 0.00000387
# # ... with 81 more rows
From your plot, though, it looks as if you want to retain all of the intermediate columns as well, which can be done this way:
foo <- function(x, ...) {
funs <- list(...)
if (is.null(names(funs))) names(funs) <- paste0("V", seq_along(funs))
out <- lapply(funs, function(f) f(x))
out$y <- do.call(mapply, c(list(FUN = prod), out))
data.frame(out)
}
tibble(myx = x) %>%
mutate(foo(myx, A1=A, B1=B, C1=C))
# # A tibble: 91 x 5
# myx A1 B1 C1 y
# <dbl> <dbl> <dbl> <dbl> <dbl>
# 1 1 0.0540 0.00000149 0.176 0.0000000141
# 2 1.1 0.0656 0.00000244 0.180 0.0000000288
# 3 1.2 0.0790 0.00000396 0.184 0.0000000576
# 4 1.3 0.0940 0.00000637 0.188 0.000000112
# 5 1.4 0.111 0.0000101 0.191 0.000000214
# 6 1.5 0.130 0.0000160 0.193 0.000000400
# 7 1.6 0.150 0.0000249 0.196 0.000000730
# 8 1.7 0.171 0.0000385 0.197 0.00000130
# 9 1.8 0.194 0.0000589 0.198 0.00000227
# 10 1.9 0.218 0.0000893 0.199 0.00000387
# # ... with 81 more rows
### or without A1= names:
tibble(myx = x) %>%
mutate(foo(myx, A, B, C))
# # A tibble: 91 x 5
# myx V1 V2 V3 y
# <dbl> <dbl> <dbl> <dbl> <dbl>
# 1 1 0.0540 0.00000149 0.176 0.0000000141
# ...
