I have a data frame where columns have strings of text. I want to search within the text of multiple columns and count the occurrence of specific words, and then make a table that reports the co-occurence (by multiple columns).
Here's a small data frame to make this reproducible:
#create dataframe
TI <- c('Studies of pancreatic cancer',
'colon, breast, and pancreatic cancer',
'cancer and CVD paper',
'CVD paper'
)
AB <- c('Autoimmune pancreatitis (AIP) is now considered a disease to be treated by diet.',
'dietary patterns, positive associations were found between pancreatic cancer risk ',
'Cardiovascular diseases (CVD) is linked to Diet',
'making this match with pancreas'
)
df <- data.frame(TI, AB)
#TI = title (of a paper)
#AB = abstract of the same paper
I then want to see how many titles have either the word 'cancer' or 'CVD' in the title (TI column), and the co-occurrence with the words 'pancreatic (or some variation)' or 'diet' in the abstract (AB column)
I can count the single occurence of the words I want with grep
pancreatic.ab <- length(grep("pancreat*", df$AB, ignore.case = TRUE, perl = FALSE))
pancreatic.ab
diet.ab <- length(grep("diet*", df$AB, ignore.case = TRUE, perl = FALSE))
diet.ab
cancer.ti <- length(grep("cancer*", df$TI, ignore.case = TRUE, perl = FALSE))
cancer.ti
CVD.ti <- length(grep("CVD", df$TI, ignore.case = TRUE, perl = FALSE))
CVD.ti
but not sure how to do this for a complicated cross tabs table.
any suggestions?
An example of desired output would be something like this
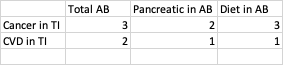
CodePudding user response:
You can bind the rows that have your targets in the TI columns, providing a label for the columns belonging to each (in my example ti). Then, grouping by that label (i.e. group_by(ti)), you can create the columns with the counts, using summarize()
bind_rows(
df %>% filter(grepl("cancer*", TI, ignore.case=T)) %>% mutate(ti="CANCER"),
df %>% filter(grepl("CVD*", TI, ignore.case=T)) %>% mutate(ti="CVD")
) %>%
group_by(ti) %>%
summarize(
TotalAB = n(),
Pancreatic = sum(grepl("pancreat*", AB, ignore.case=T)),
Diet = sum(grepl("diet*", AB, ignore.case=T))
)
Output:
ti TotalAB Pancreatic Diet
<chr> <int> <int> <int>
1 CANCER 3 2 3
2 CVD 2 1 1
