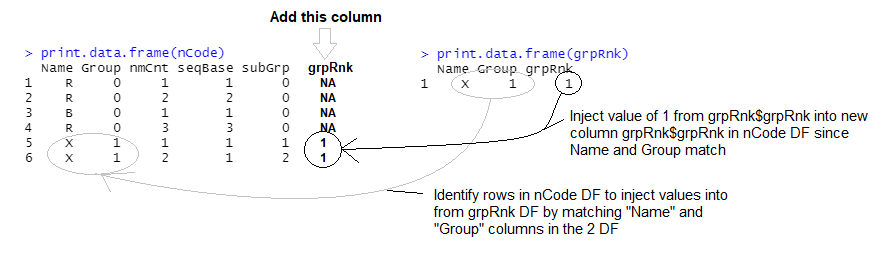
I am trying to match these two data frames, nCode and grpRnk, as illustrated below.
Using the code shown at the bottom, I've been able to get the below output whereby the last column on the right shows the correct values, but I don't need those extra columns from grpRnk$Name through $subGrp (columns 6 -10) and I had specified a name for the last column of grpRnk but I instead get $grpRnk. Am I using this merge() function correctly? Is there a more correct or efficient way to do this sort of multi-factor matching?
Name Group nmCnt seqBase subGrp grpRnk$Name $Group $nmCnt $seqBase $subGrp $grpRnk
<chr> <dbl> <int> <int> <int> <chr> <dbl> <int> <int> <int> <int>
1 R 0 1 1 0 B 0 1 1 0 NA
2 R 0 2 2 0 R 0 1 1 0 NA
3 B 0 1 1 0 R 0 2 2 0 NA
4 R 0 3 3 0 R 0 3 3 0 NA
5 X 1 1 1 1 X 1 1 1 1 1
6 X 1 2 1 2 X 1 2 1 2 1
The code:
library(dplyr)
myDF1 <-
data.frame(
Name = c("R","R","B","R","X","X"),
Group = c(0,0,0,0,1,1)
)
nCode <- myDF1 %>%
group_by(Name) %>%
mutate(nmCnt = row_number()) %>%
ungroup() %>%
mutate(seqBase = ifelse(Group == 0 | Group != lag(Group), nmCnt,0)) %>%
mutate(seqBase = na_if(seqBase, 0)) %>%
group_by(Name) %>%
fill(seqBase) %>%
mutate(seqBase = match(seqBase, unique(seqBase))) %>%
ungroup() %>%
mutate(subGrp = as.integer(ifelse(Group > 0, sapply(1:n(), function(x) sum(Name[1:x]==Name[x] & Group[1:x] == Group[x])),0)))
grpRnk <- nCode %>% select(Name,Group,nmCnt) %>%
filter(Group > 0) %>%
group_by(Name,Group) %>%
slice(which.min(Group)) %>%
ungroup() %>%
arrange(nmCnt) %>%
mutate(grpRnk = dense_rank(nmCnt)) %>%
select (-nmCnt)
nCode %>% mutate(grpRnk = merge(nCode,grpRnk, by=c("Name","Group"), all.x=T))
CodePudding user response:
You need to specify what column you want to extract into your newly created grpRnk column. You can achieve this by adding $grpRnk to the end of your merge() statement.
Here I have first a solution with merge():
library(tidyverse)
myDF1 <-
data.frame(
Name = c("R","R","B","R","X","X"),
Group = c(0,0,0,0,1,1)
)
nCode <- myDF1 %>%
group_by(Name) %>%
mutate(nmCnt = row_number()) %>%
ungroup() %>%
mutate(seqBase = ifelse(Group == 0 | Group != lag(Group), nmCnt,0)) %>%
mutate(seqBase = na_if(seqBase, 0)) %>%
group_by(Name) %>%
fill(seqBase) %>%
mutate(seqBase = match(seqBase, unique(seqBase))) %>%
ungroup() %>%
mutate(subGrp = as.integer(ifelse(Group > 0, sapply(1:n(), function(x) sum(Name[1:x]==Name[x] & Group[1:x] == Group[x])),0)))
grpRnk <- nCode %>% select(Name,Group,nmCnt) %>%
filter(Group > 0) %>%
group_by(Name,Group) %>%
slice(which.min(Group)) %>%
ungroup() %>%
arrange(nmCnt) %>%
mutate(grpRnk = dense_rank(nmCnt)) %>%
select (-nmCnt)
nCode %>% mutate(grpRnk = merge(nCode,grpRnk, by=c("Name","Group"), all.x = TRUE)$grpRnk)
#> # A tibble: 6 × 6
#> Name Group nmCnt seqBase subGrp grpRnk
#> <chr> <dbl> <int> <int> <int> <int>
#> 1 R 0 1 1 0 NA
#> 2 R 0 2 2 0 NA
#> 3 B 0 1 1 0 NA
#> 4 R 0 3 3 0 NA
#> 5 X 1 1 1 1 1
#> 6 X 1 2 1 2 1
However, you can achieve the same result using only merge() like this:
merge(nCode,grpRnk, by=c("Name","Group"), all.x = TRUE)
#> Name Group nmCnt seqBase subGrp grpRnk
#> 1 B 0 1 1 0 NA
#> 2 R 0 1 1 0 NA
#> 3 R 0 2 2 0 NA
#> 4 R 0 3 3 0 NA
#> 5 X 1 1 1 1 1
#> 6 X 1 2 1 2 1
And here is another solution using left_join() from the dplyr package:
left_join(nCode, grpRnk, by = c("Name", "Group"))
#> # A tibble: 6 × 6
#> Name Group nmCnt seqBase subGrp grpRnk
#> <chr> <dbl> <int> <int> <int> <int>
#> 1 R 0 1 1 0 NA
#> 2 R 0 2 2 0 NA
#> 3 B 0 1 1 0 NA
#> 4 R 0 3 3 0 NA
#> 5 X 1 1 1 1 1
#> 6 X 1 2 1 2 1
Created on 2022-09-19 with reprex v2.0.2