I have this code, it produces a random matrix of 1s and 0s. I want to create a graph from this matrix where the 1s in the matrix represent a node and each node has a maximum of 3 edges. How can i implement this, please help?
import numpy as np
from random import sample
N = int(input("Enter the number of nodes:"))
my_matrix = np.zeros((N,N), dtype='int8')
rows = sample(range(N), N)
cols = sample(range(N), N)
points = zip(rows, cols)
for x, y in points:
my_matrix[x, y] = 1
print(my_matrix)
CodePudding user response:
If you matrix is just random, probably, you don't need it. Instead, you can create graph from list of edges
import networkx as nx
from random import sample
import numpy as np
from numpy.random import randint
n = 7 # number of nodes in graph
max_connections = int(input("Enter max connections per node:")) # input: 3
nodes = np.arange(n)
# create graph based on list of edges [(0, 1), (0, 4), ...]
gr = nx.Graph([
# for each node select <= 'max_connections' nodes as connections
(i, j) for i in range(n) for j in sample(nodes[nodes != i].tolist(), randint(1, max_connections 1))
])
# check number of connections
for n in gr.nodes():
nei = list(gr.neighbors(n))
while len(nei) > max_connections:
gr.remove_edge(n, random.choice(nei))
nei = list(gr.neighbors(n))
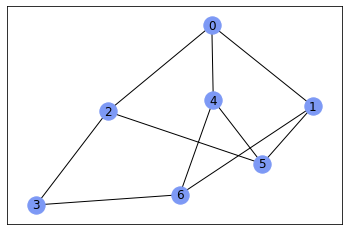
nx.draw_networkx(gr, with_labels=True, node_color='#7d99f5')
And you can get adjacency matrix using nx.adjacency_matrix()
nx.adjacency_matrix(gr, nodelist=sorted(gr.nodes())).todense()
matrix([[0, 1, 1, 0, 1, 0, 0],
[1, 0, 0, 0, 0, 1, 1],
[1, 0, 0, 1, 0, 1, 0],
[0, 0, 1, 0, 0, 0, 1],
[1, 0, 0, 0, 0, 1, 1],
[0, 1, 1, 0, 1, 0, 0],
[0, 1, 0, 1, 1, 0, 0]])
CodePudding user response:
Addition to your code (function check_graph () fix two problems we have discussed). Also, according your way of generation of adjacency matrix, you won't face second problem, so you can comment out second section in function.
def check_graph(graph, max_conn):
# 1) remove self loops
graph.remove_edges_from(nx.selfloop_edges(graph))
# 2) remove random edge(s) if limit of edges per node have been exceeded
for i in graph.nodes():
# list of connections - nodes that are connected to the selected node 'i'
nei = list(graph.neighbors(i))
if len(nei) > max_conn:
graph.remove_edges_from(
# like if len(nei) - max_conn = 5 - 4 = 1, then one random edge will be selected
np.random.choice(nei, size=(len(nei)-max_conn))
)
# <-- insert your code here --> N = 20
gr = nx.from_numpy_matrix(my_matrix)
check_graph(gr, max_conn=N)
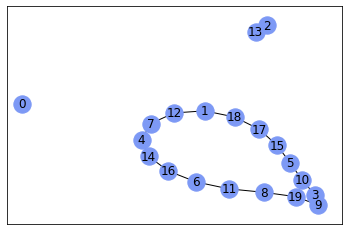
nx.draw_networkx(gr, with_labels=True, node_color='#7d99f5')
Result looks a bit strange for me, but I don't know purposes of your graph, probably, it's okay.