#generating 100 uniformly distributed numbers
u1 <- runif(100,0,1)
u2 <- runif(100,0,1)
x1 <- function(x, y) {
return(sqrt(-2 * log(x) * cos(2 * pi * y)))
}
x2 <- function(x, y) {
return(sqrt(-2 * log(x) * sin(2 * pi * y)))
}
#applying x1
x1_vals <- mapply(x1, u1, u2)
#applying x2
x2_vals <- mapply(x2, u1, u2)
Hi I want to write a box muller function, this is part of my attempt above, I'm trying to avoid using loops as much as possible(for loops especially) However, I keep getting NA values for my x1/x2 functions. I can't figure out whats wrong.
CodePudding user response:
Those functions are already vectorized. Don't need to wrap mapply around them. Just give the two vectors to x1 and x2
x1(u1,u2)
[1] NaN NaN NaN NaN 0.3088330
[6] NaN 0.7866889 NaN NaN 1.7102801
[11] 2.1886770 NaN 1.5473627 1.0644378 0.8499208
snipped remaining 100 values
Vectorization is a feature of the R language. If the expressions in the function body can all take vectors and return vectors of equal length then no loop wrapper is needed.
You are getting NA's because the domain of the arguments to sin and cos are causing both positive and negative values to be given to sqrt.
CodePudding user response:
Here is the Box-Muller formula corrected.
x1 <- function(x, y) sqrt(-2 * log(x)) * cos(2 * pi * y)
x2 <- function(x, y) sqrt(-2 * log(x)) * sin(2 * pi * y)
u1 <- runif(1000,0,1)
u2 <- runif(1000,0,1)
# applying x1
x1_vals <- x1(u1, u2)
all(is.finite(x1_vals))
#> [1] TRUE
# applying x2
x2_vals <- x2(u1, u2)
all(is.finite(x2_vals))
#> [1] TRUE
old_par <- par(mfrow = c(1, 2))
hist(x1_vals, freq = FALSE)
curve(dnorm, from = -4, to = 4, add = TRUE)
hist(x2_vals, freq = FALSE)
curve(dnorm, from = -4, to = 4, add = TRUE)
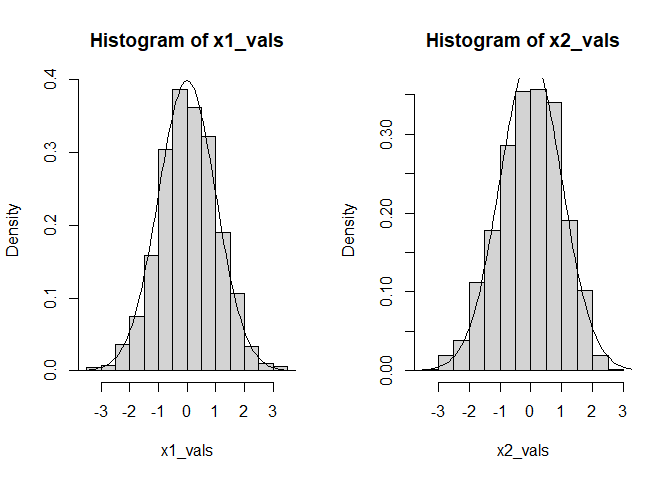
par(old_par)
Created on 2022-09-28 with reprex v2.0.2
