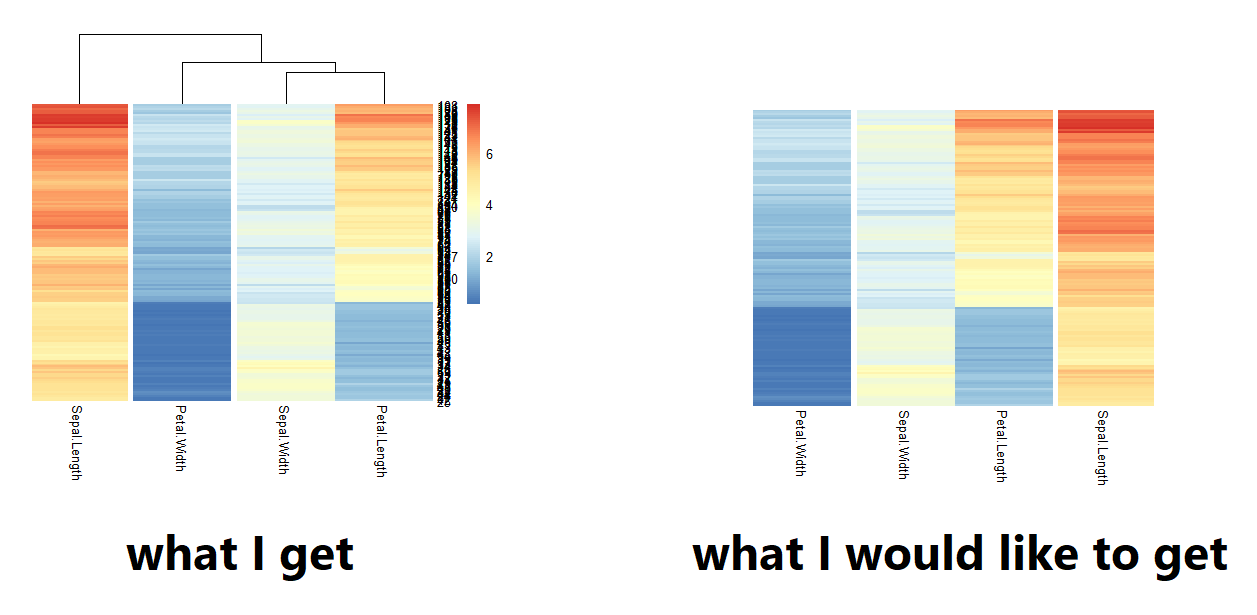
I have created a heatmap with a corresponding dendogram based on hierarchical clustering with {pheatmap}. I would like to change the order of the leaves in the dendogram, manually, based on what I see visually.
First, can anyone confirm that this is statistically correct and allowed? (in theory that should not change the between-cluster distance, but maybe I am wrong).
Second, any suggestions on how to change the order of the leaves would be appreciated!
A reproductible example with the iris data:
data(iris)
pheatmap(iris[1:4], cutree_cols = 3)
CodePudding user response:
For your example you can use a callback function to reorder the columns, e.g.
library(pheatmap)
data(iris)
colnames(iris)
#> [1] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width" "Species"
callback = function(hc, mat){
sv = svd(t(mat))$v[,c(1)]
dend = reorder(as.dendrogram(hc), wts = sv^2)
as.hclust(dend)
}
#svd(t(iris[c(4, 2, 3, 1)]))$v[,1]
pheatmap(iris[c(4, 2, 3, 1)], cutree_cols = 3, clustering_callback = callback)
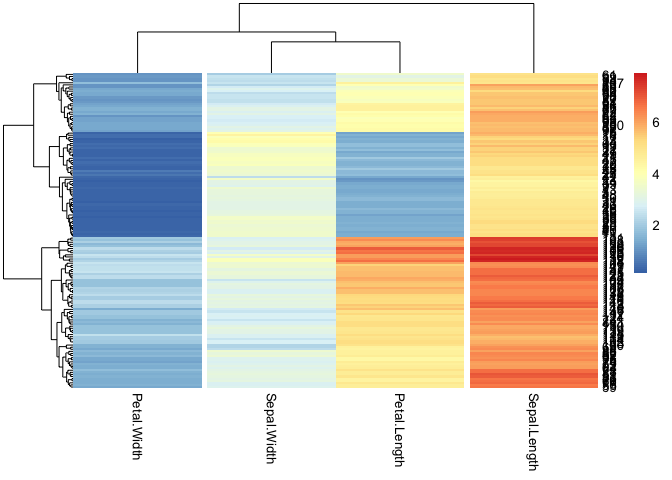
Created on 2022-09-28 by the reprex package (v2.0.1)
For your actual data, you will probably need to fiddle around with the weights to get the columns in your desired order, e.g.
library(pheatmap)
data(iris)
colnames(iris)
#> [1] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width" "Species"
callback = function(hc, mat){
sv = svd(t(mat))$v[,c(2)]
dend = reorder(as.dendrogram(hc), wts = sv)
as.hclust(dend)
}
#svd(t(iris[c(4, 2, 3, 1)]))$v[,2]
pheatmap(iris[c(4, 2, 3, 1)], cutree_cols = 3, clustering_callback = callback)
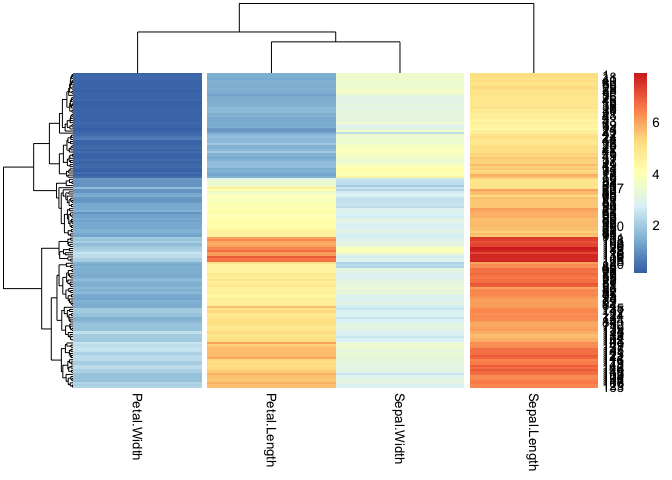
Created on 2022-09-28 by the reprex package (v2.0.1)
This feature is described briefly at the end of the help file:
?pheatmap
...
# Modify ordering of the clusters using clustering callback option
callback = function(hc, mat){
sv = svd(t(mat))$v[,1]
dend = reorder(as.dendrogram(hc), wts = sv)
as.hclust(dend)
}
pheatmap(test, clustering_callback = callback)
## Not run:
# Same using dendsort package
library(dendsort)
callback = function(hc, ...){dendsort(hc)}
pheatmap(test, clustering_callback = callback)
## End(Not run)
CodePudding user response:
To achieve the desired output in your example, you can add cluster_cols=F, reorder the columns manually, and add gaps_col to specify the gaps manually:
data(iris)
pheatmap::pheatmap(
iris[c(4,2,3,1)],
cluster_cols=F,
cluster_rows=F,
gaps_col=c(1,3)
)
You can also use reorder.hclust from vegan to reorder the branches of the clustering tree without having to convert the hclust object to a dendrogram and back. Often a good weight for reordering the branches is the first dimension in a PCA of the input (or MDS if the input is a distance matrix):
data(iris)
df=iris[1:4]
library(vegan) # for reorder.hclust
hc=reorder(hclust(dist(t(scale(df)))),prcomp(t(scale(df)))$x[,1])
# hc=reorder(hclust(as.dist(df)),cmdscale(df)[,1]) # for distance matrix
pheatmap::pheatmap(
df,
cluster_rows=F,
clustering_callback=\(...)hc,
cutree_cols=3
)