I would like to create an image file containing the color key for an R plot that uses a colorbrewer palette. This needs to be a separate file from the plot itself. Colorbrewer produces a nice plot describing the palette:
display.brewer.pal(11, "RdBu")
However, I cannot find any way to alter this plot -- e.g. add values, a title, control size, and dimensions, and save it as an image file. Is there a way to do this with the output of the display.brewer.pal function? Or some alternative using the underlying palette?
I am looking for a completely separate legend based on the palette, not a legend produced by a plot function.
CodePudding user response:
I think you need to modify the display.brewer.pal function, but it's pretty simple, so this isn't hard. For example:
mypalette <- function (n, name, xlab = paste(name, palattr),
transpose = FALSE, ...)
{
# Modified version of RColorBrewer::display.brewer.pal
if (!(name %in% namelist)) {
stop(paste(name, "is not a valid palette name for brewer.pal\n"))
}
if (n < 3) {
warning("minimal value for n is 3, displaying requested palette with 3 different levels\n")
return(display.brewer.pal(3, name))
}
if (n > maxcolors[which(name == namelist)]) {
warning(paste("n too large, allowed maximum for palette",
name, "is", maxcolors[which(name == namelist)]),
"\nDisplaying the palette you asked for with that many colors\n")
return(display.brewer.pal(maxcolors[which(name == namelist)],
name))
}
if (length(which(name == quallist)) > 0)
palattr <- "(qualitative)"
if (length(which(name == divlist)) > 0)
palattr <- "(divergent)"
if (length(which(name == seqlist)) > 0)
palattr <- "(sequential)"
if (transpose)
image(1, 1:n, t(as.matrix(1:n)), col = brewer.pal(n, name),
xlab = xlab, ylab = "", xaxt = "n", yaxt = "n",
bty = "n", ...)
else
image(1:n, 1, as.matrix(1:n), col = brewer.pal(n, name),
xlab = xlab, ylab = "", xaxt = "n", yaxt = "n",
bty = "n", ...)
}
environment(mypalette) <- environment(RColorBrewer::display.brewer.pal)
par(mar = c(1, 10, 5, 10))
mypalette(11, "RdBu", main = "My Palette", xlab = "", transpose = TRUE)
axis(4, at = 1:11, labels = LETTERS[1:11], las = 1)
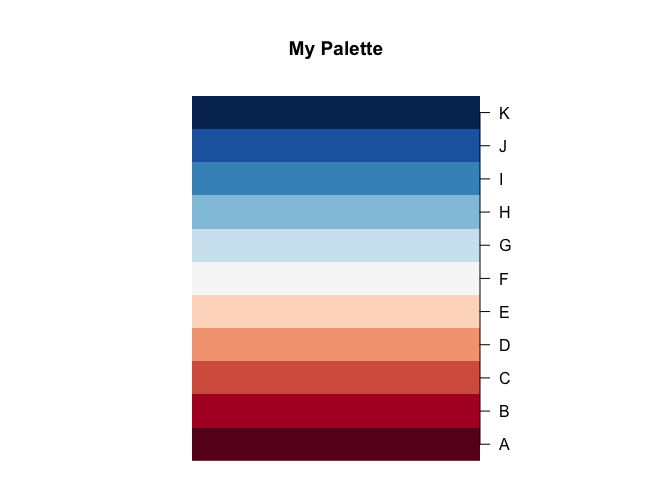
Created on 2022-10-02 with reprex v2.0.2
