I'm working on a url to get XML data and make it a dataframe. I tried the following code:
fileURL <- "https://data.ny.gov/api/views/ngbt-9rwf/rows.xml"
xData <- getURL(fileURL)
xmlfile <- xmlTreeParse(xData)
xmltop = xmlRoot(xmlfile)
plantcat <- xmlSApply(xmltop, function(x) xmlSApply(x, xmlValue))
plantcat_df <- data.frame(t(plantcat),row.names=NULL)
View(plantcat_df)
But my output is all in one row, with thousands columns. Is there any way I can break them into different columns? Here is the URL of my 
Thank you.
CodePudding user response:
Looking at the website, it uses a SODA API for their datasets. You can use RSocrata package to retrieve them by running the code install.packages("RSocrata") and then you can simply call the package and use the unique dataset key to retrieve the dataset through R.
library(RSocrata)
#list all the available dataset in the website
list.datasets <- RSocrata::ls.socrata("https://data.ny.gov")
# retrieve the data using the unique dataset key "ngbt-9rwf"
df <- RSocrata::read.socrata("https://data.ny.gov/d/ngbt-9rwf")
CodePudding user response:
Because you have used xmlSApply() at wrong node. Following code work. just only need to replace the local path with your URL.
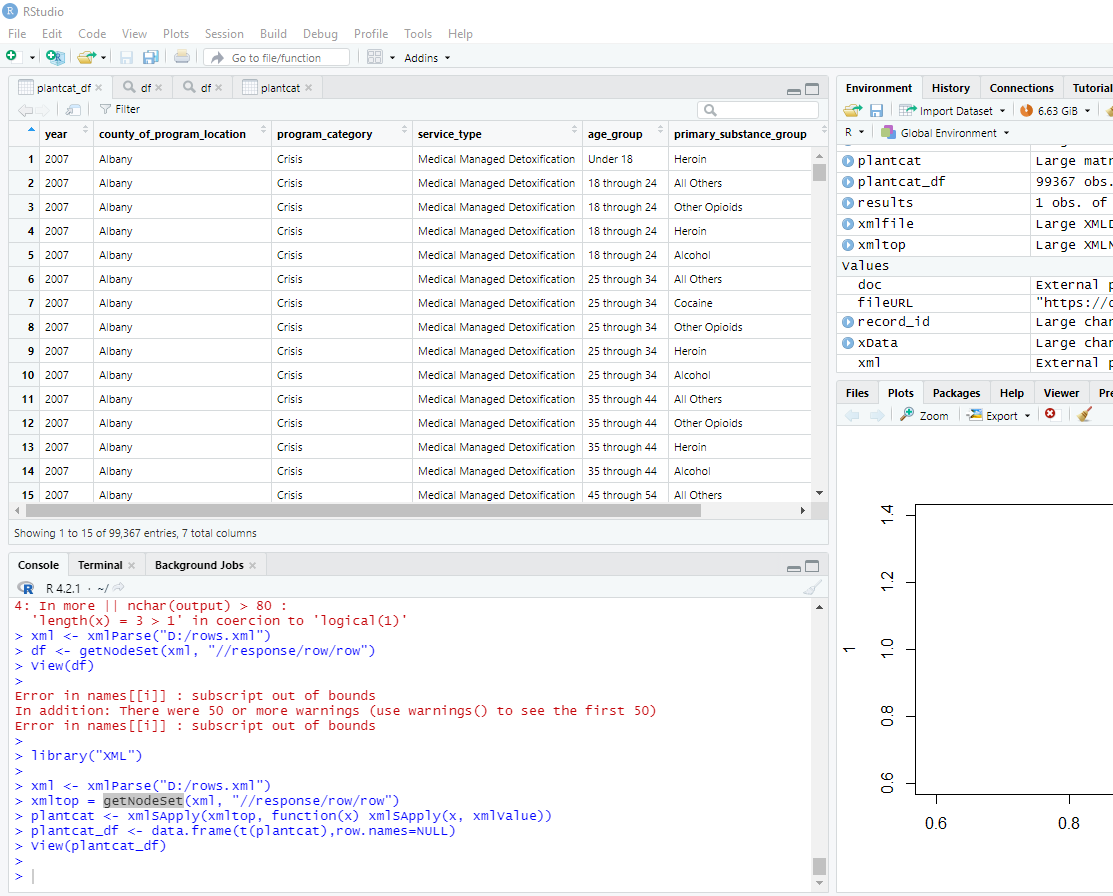
library("XML")
xml <- xmlParse("D:/rows.xml")
xmltop = getNodeSet(xml, "//response/row/row")
plantcat <- xmlSApply(xmltop, function(x) xmlSApply(x, xmlValue))
plantcat_df <- data.frame(t(plantcat),row.names=NULL)
View(plantcat_df)