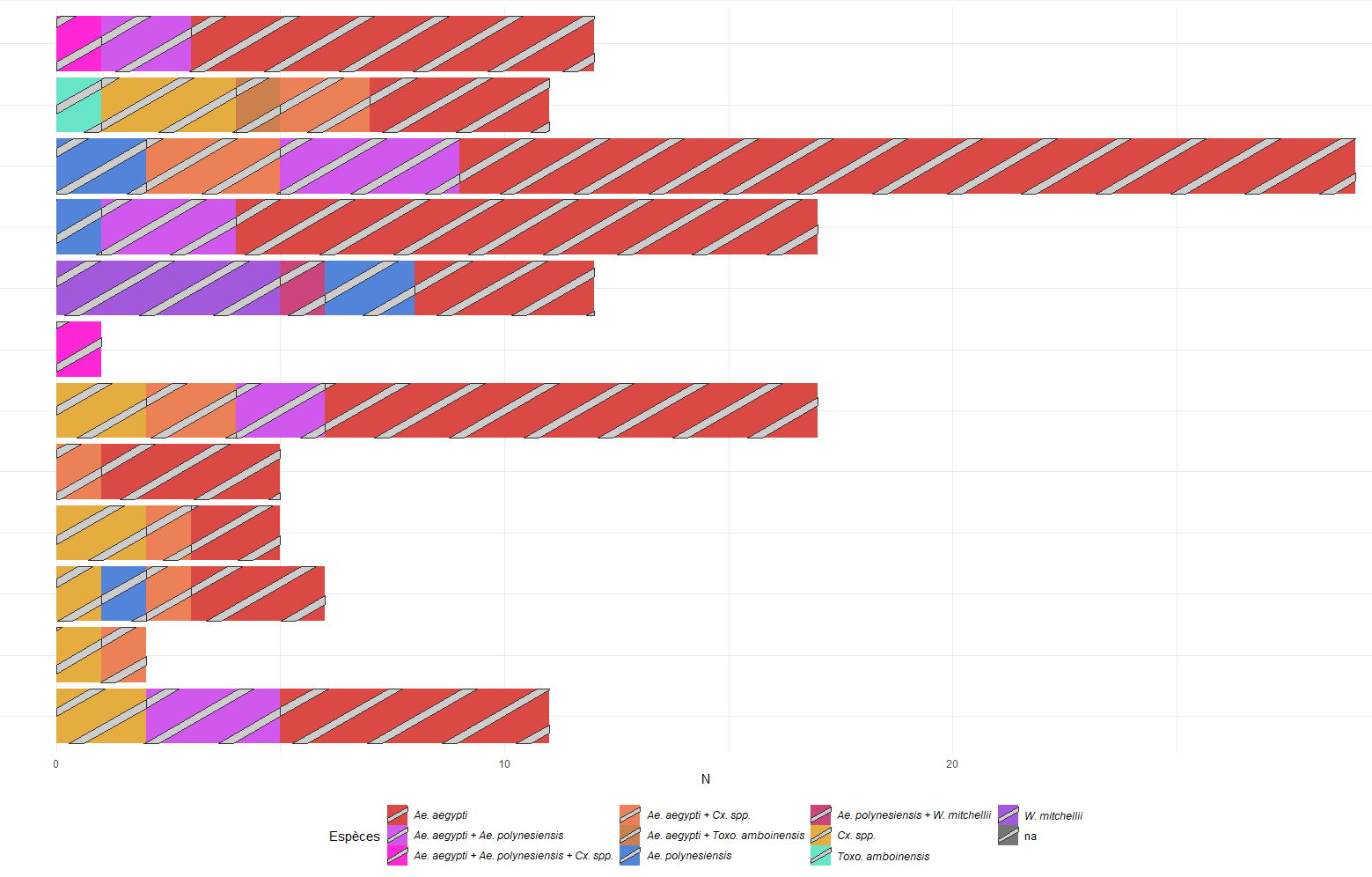
I tried to represent species presence into containers breeding site. I used a bar plot and a color code to represent my different species. I first tried to specify a color for each value of my sp variable, but it appeared to be to much colored as I have combination of multiple species in a breeding site. In order to simplify the visual of the plot, I tried to represent the presence of two species in the same container by adding a colored pattern of a species above the color of another, but did not succeed.
Here's my code below I used. I tried but did not understand the use of scale_pattern_manual
Any suggestions ?
x11(); ggppt<-Tabagg %>%
filter(!(type_gîtes %in% "na")) %>%
filter(pres_larve %in% "Oui") %>%
filter(!(sp %in% "na")) %>%
ggplot()
aes(x = type_gîtes, fill = sp)
geom_bar()
labs(x = "Type gîte",
y = "N", fill = "Espèces")
coord_flip()
theme_minimal()
theme(legend.text.align = 0,
legend.position = "bottom")
scale_fill_manual(name = "Espèces" ,
values = c("Ae. aegypti" = "#DA4943",
"Ae. aegypti Ae. polynesiensis" = "#D058EC",
"Ae. aegypti Ae. polynesiensis Cx. spp." = "#FF27D5",
"Ae. aegypti Cx. spp." = "#EC8158",
"Ae. aegypti Toxo. amboinensis" = "#CC804D",
"Ae. polynesiensis" = "#5284D9",
"Ae. polynesiensis W. mitchellii" = "#CB447C",
"Cx. spp." = "#E5AD3F",
"Toxo. amboinensis" = "#67E5C8",
"W. mitchellii" = "#A259DB",
"na" = "#757575"
),
labels = c(expression(italic("Ae. aegypti"),
italic("Ae. aegypti Ae. polynesiensis"),
italic("Ae. aegypti Ae. polynesiensis Cx. spp."),
italic("Ae. aegypti Cx. spp."),
italic("Ae. aegypti Toxo. amboinensis"),
italic("Ae. polynesiensis"),
italic("Ae. polynesiensis W. mitchellii"),
italic("Cx. spp."),
italic("Toxo. amboinensis"),
italic("W. mitchellii"),
"na")))
geom_bar_pattern()
scale_pattern_manual(values=c("Ae. aegypti Ae. polynesiensis" ="Stripe")); ggppt
Here's the plot generated
CodePudding user response:
It looks like you're new to SO; welcome to the community! If you want great answers quickly, it's best to make your question reproducible. This includes sample data like the output from dput() or reprex::reprex() and any libraries you are using. Check it out: 
CodePudding user response:
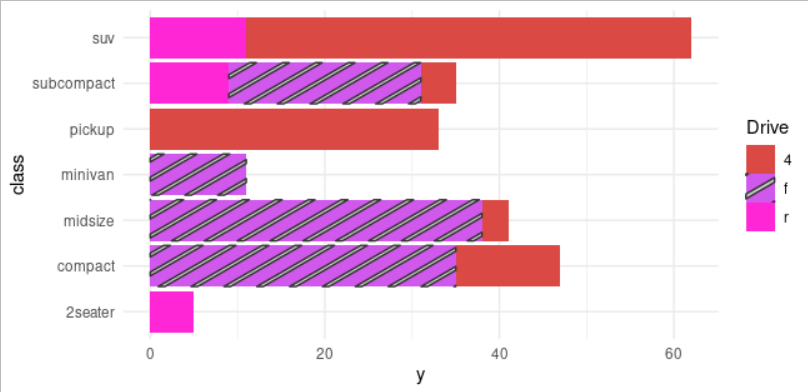
You may want to try something like this:
Data
library(tidyverse)
library(ggpattern)
dat <- data.frame(drv = c("4", "4", "4", "4", "4", "f", "f", "f", "f", "r", "r", "r"),
class = c("compact", "midsize", "pickup", "subcompact", "suv",
"compact", "midsize", "minivan", "subcompact",
"2seater", "subcompact", "suv"),
y = c(12L, 3L, 33L, 4L, 51L, 35L, 38L, 11L, 22L, 5L, 9L, 11L))
dat
Give the same name for scale_fill_manual() and scale_pattern_manual(). Also, specify the group (or groups) you would like to show the pattern (or patterns).
dat %>% ggplot(aes(x = class, y = y, fill = drv, pattern = drv))
# geom_col() # not necessary
geom_col_pattern()
coord_flip()
theme_minimal()
scale_fill_manual(
name = "Drive",
values = c(
"4" = "#DA4943",
"f" = "#D058EC",
"r" = "#FF27D5"
)
)
scale_pattern_manual(
name = "Drive",
values = c(
"4" = "none",
"f" = "stripe",
"r" = "none"
)
)