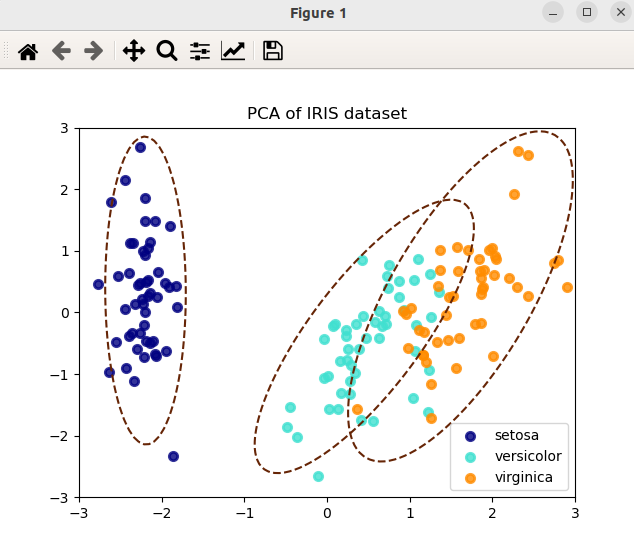
I am working creating a PCA combined with a Mahalanobis distance function to remove any outliers detected from the PCA transformation. I have come across this article which uses R: 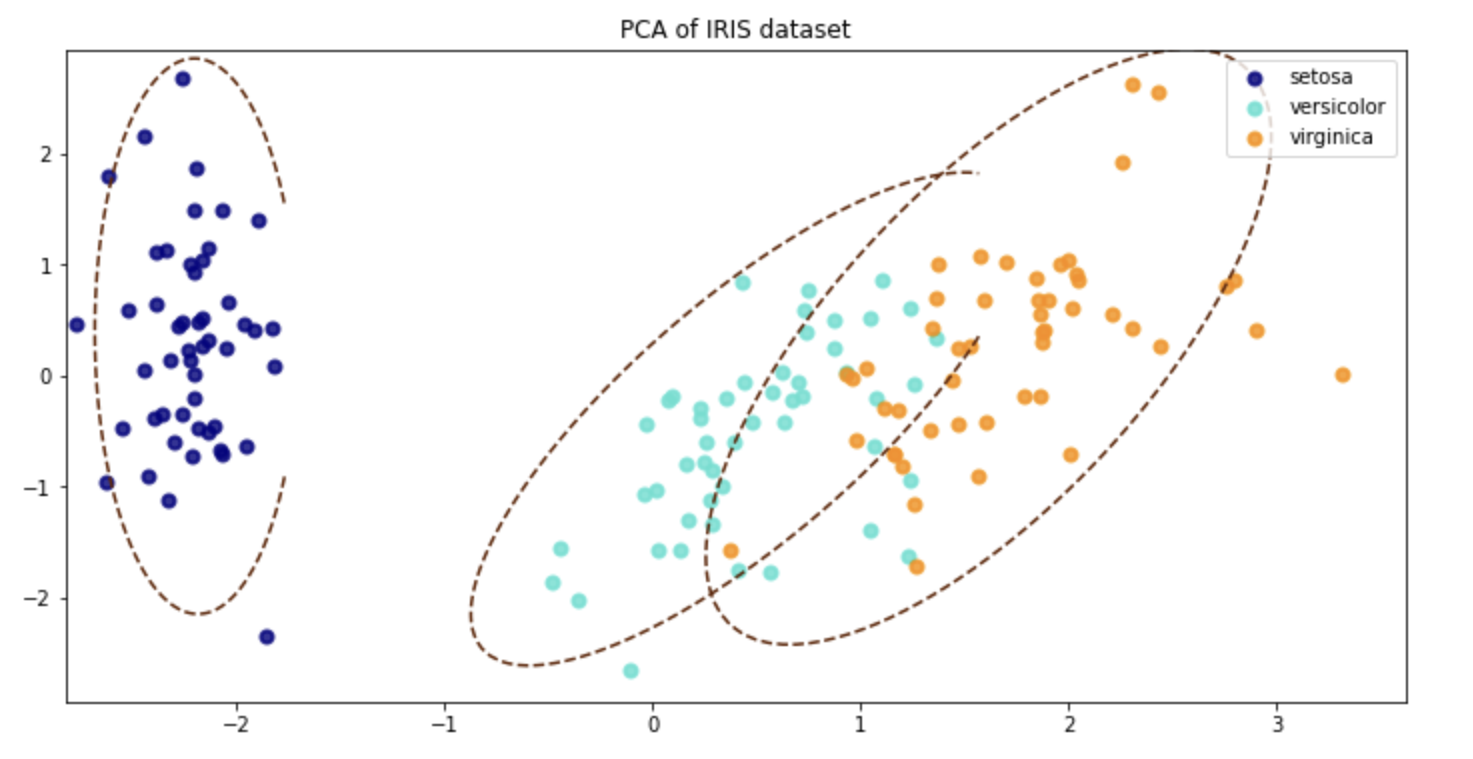
CodePudding user response:
Please add plt.xlim() and plt.ylim():
plt.legend(loc="best", shadow=False, scatterpoints=1)
plt.title("PCA of IRIS dataset")
plt.xlim(-3,4) # ADD
plt.ylim(-3,3) # ADD
xx, yy = np.meshgrid(
np.linspace(plt.xlim()[0], plt.xlim()[1], 100),
np.linspace(plt.ylim()[0], plt.ylim()[1], 100),)
zz = np.c_[xx.ravel(), yy.ravel()]